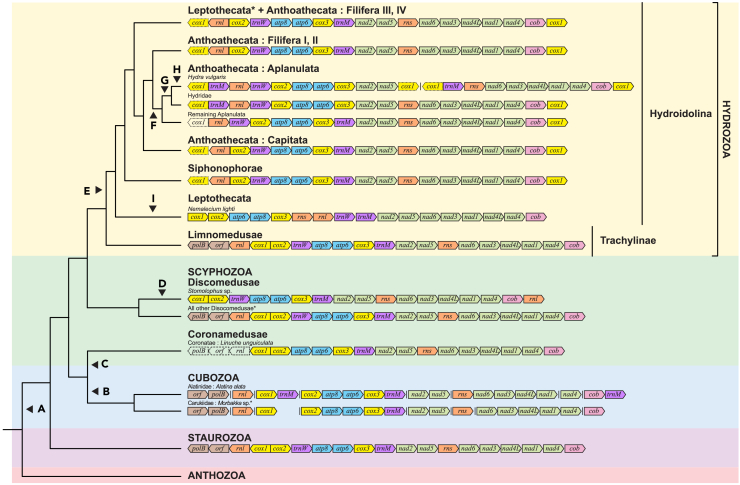
Figure 3.
Comparison of gene order of Medusozoa mitochondrial genes with reference to the Maximum Likelihood (ML) phylogeny, using complete cnidarian mitogenomes
For mitogenomes with multiple chromosomes, vertical lines denote breaks between each chromosomal fragment. Complete mitochondrial genes are indicated in color, while pseudogenes (as indicated on GenBank) are colored but outlined with dotted lines. Missing genes are represented by uncolored boxes outlined with dotted lines.
(A–I) Mitochondrial rearrangements indicated using black arrows: (A) Ancestral medusozoan gene order (AMGO); (B) fragmentation of mitochondrial genome, loss of trnW; (C) loss of trnW, rnl, polB, and orf314, (D) inverse transposition of rnl and loss of polB and orf314; (E) loss of gene pair polB-orf314, and duplication, transposition, and inversion of cox1; (F) inversion of rnl and transposition of trnW; (G) transposition of trnM; (H) fragmentation and duplication of cox1; (I) inversion and transposition of rnl, as well as transposition of rns, cox1, atp6, and nad1. Asterisks (∗) denote the phylogenetic positions of specimens assembled in current study: Scyphozoa – Acromitus sp., Cassiopea sp., Phyllorhiza punctata; Hydrozoa – Zygocanna sp., Aequorea sp.; Cubozoa – Morbakka sp.