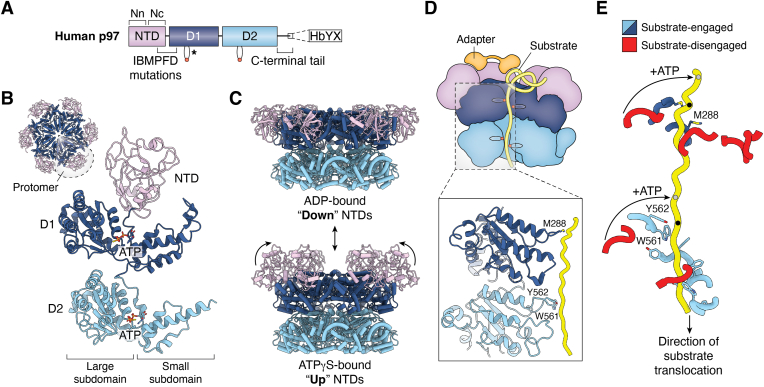
Figure 2.
Structure and substrate threading mechanism of p97.A, domain schematic of human p97 colored by domain (NTD: purple, D1: dark blue, and D2: light blue). Location of pore loops in D1 and D2 are shown; degenerate residues in D1 are indicated (∗). The two lobes of the NTD (N- and C-terminal lobes, Nn and Nc) are indicated, as is the location of most IBMPFD mutations, namely the NTD-D1 interface. The HbYX motif at the end of the C-terminal tail is also shown. B, protomer of ATPγS-bound p97 from an intact hexamer (PDB 5FTN) (52), colored as in (A). A downscaled top view of the full hexamer is also shown, with one protomer circled to delineate protomer boundaries. C, side views of ADP-bound (top, PDB 5FTK) and ATPγS-bound (bottom, PDB 5FTN) p97 hexamers (52), showing rotation and elevation of the NTDs above the D1 ring in the ATPγS-bound state. D, illustration of adapter-mediated substrate threading through the p97 central pore, with D1 and D2 pore loops shown. The adapter is colored in orange, and the substrate in yellow. Below, an enlarged view of pore loop contacts is shown (PDB 6OA9) (23). E, view of substrate in the Cdc48 channel (PDB 6OA9) (23), showing a spiral arrangement of pore loops in D1 and D2. Pore loops engaged with substrate are shown in blue; those not engaged are in red. The highest substrate contacts in D1 and D2 are marked by black dots; hydrolysis and subsequent ATP binding by the disengaged protomers is proposed to drive translocation by two amino acid steps whereby conformational changes enable pore loop engagement with the next site along the substrate (gray dots). IBMPFD, inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia; NTD, N-terminal domain; PDB, Protein Data Bank.