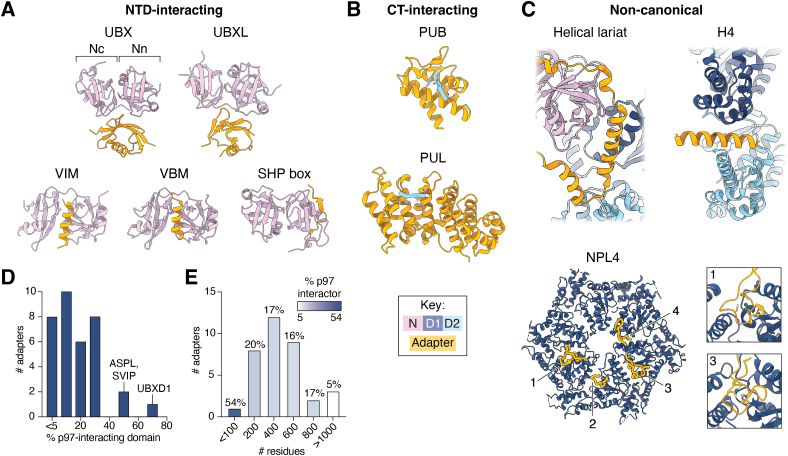
Figure 3.
Architecture and interaction modes of p97 adapters. Structures of individual p97-interacting domains bound to the p97 NTD, CT tail, or other sites are shown, highlighting the diversity of adapter interaction modes. A, structures of the p97 NTD bound to NTD-interacting domains. The p97 NTD lobes are indicated in the UBX-bound structure. PDB IDs: UBX: 5X4L (63), UBXL: 4KDI (66), VIM: 3TIW (67), VBM: 5EPP (68), and SHP box: 5GLF (69). B, structures of the HbYX motif at the end of the p97 CT tail bound to CT-interacting domains. PDB IDs: PUB: 2HPL (73) and PUL: 3EBB (71). C, structures of noncanonical p97-interacting domains bound to p97 or a homolog thereof. PDB IDs: helical lariat: 8FCL (24), H4: 8FCR (24), and NPL4: 6OA9 (23). The four interacting motifs in NPL4 are numbered counterclockwise: zinc finger 1 (ZF1, 1), N-terminal bundle (NTB, 2), zinc finger 2 (ZF2, 3), and β-strand finger (4). Enlarged views of ZF1 and ZF2 are shown at right; Zn2+ is colored in teal. D, histogram of the proportion of adapters corresponding to p97-interacting domains (determined by % of total sequence length). E, histogram of adapter length. Bars are colored by average % p97 interactor of adapters in that bin (determined by % of total sequence length). The color key indicates p97 domains, as in Figure 2, and adapters. CT, C-terminal; NTD, N-terminal domain; PDB, Protein Data Bank; PUB, peptide:N-glycanase and UBA or UBX-containing proteins; PUL, PLAP, Ufd3p, and Lub1p; UBX, ubiquitin regulatory X; UBXL, UBX-like; VBM, VCP-binding motif; VIM, VCP-interacting motif.