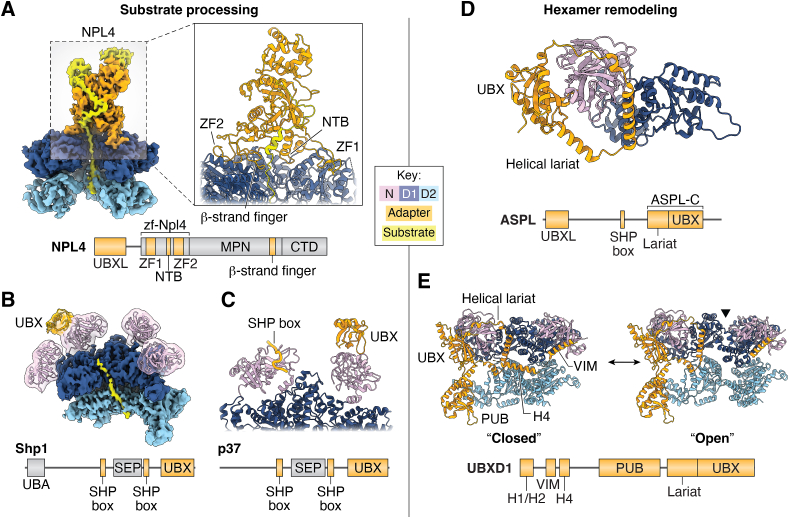
Figure 4.
Structures of intact p97-adapter complexes. Structures of p97-adapter complexes involved in substrate processing (left) or hexamer remodeling (right). Domain colors are indicated by the key. Domain schematics (not to scale) of each adapter are shown below the corresponding structure, with p97-interacting domains colored in orange and other domains in gray. A, (left) cryo-EM structure of yeast Cdc48 bound to the UFD1/NPL4 adapter (PDB 6OA9) (23), unfolding an ubiquitylated substrate and threading it through the Cdc48 central channel. (Right) Enlarged view of contacts made between NPL4 and Cdc48. B, cryo-EM structure of Cdc48 bound to substrate and the Shp1 adapter (PDB 6OPC) (56). A filtered transparent map and model of the NTDs and UBX is shown over the sharpened map of the D1, D2, and substrate. C, cryo-EM structure of a SHP box and UBX domain of p37 bound to adjacent NTDs of an actively processing p97 complex (PDB 8B5R) (43). D, crystal structure of an ASPL truncation construct containing the UBX and helical lariat domains (ASPL-C) bound to a human p97 construct (PDB 5IFS) (74). E, cryo-EM structures of human p97 bound to UBXD1 (closed: PDB 8FCR, open: PDB 8FCM) (24), showing separation of adjacent p97 protomers coordinated by multiple UBXD1 contacts. The hexamer seam is indicated in the open state by a black triangle. NTB, N-terminal bundle; NTD, N-terminal domain; PDB, Protein Data Bank; UBX, ubiquitin regulatory X; ZF, zinc finger.