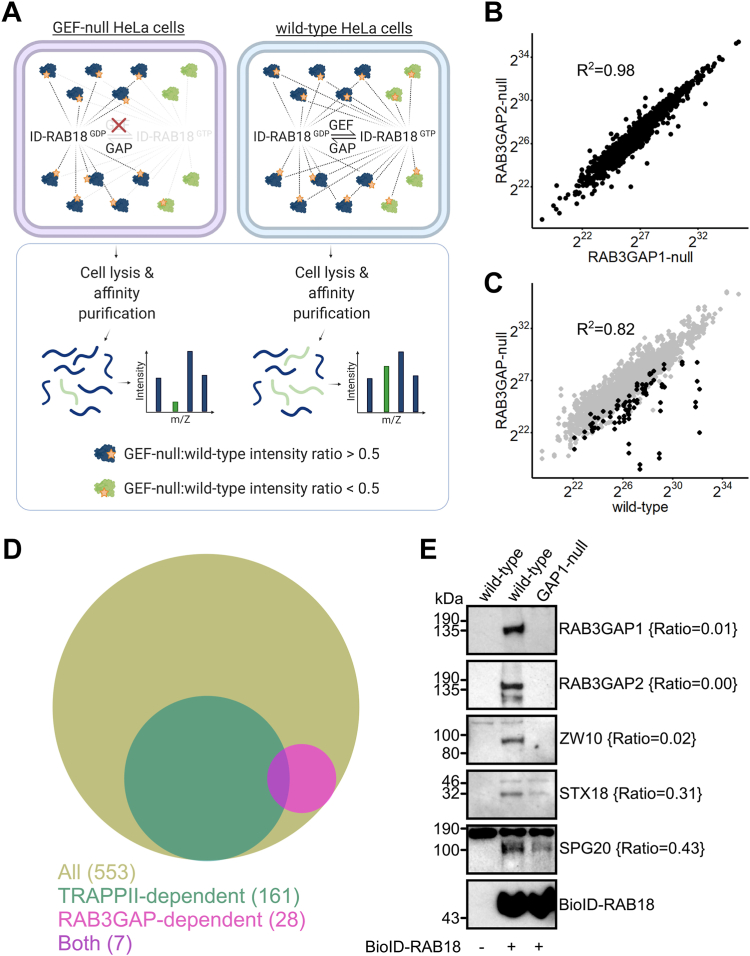
Figure 1.
RAB3GAP-dependent RAB18 interactions in HeLa cells.A, schematic to show experimental approach. Proximity biotinylation of guanine nucleotide exchange factor (GEF)-dependent interactors by BirA∗-RAB18 (ID-RAB18) is disrupted in GEF-null cells. GEF-independent interactors are biotinylated in both GEF-null and WT cells. Following affinity purification, GEF-dependent interactions are determined by label-free quantitative (LFQ) intensity ratios. B, plot to show correlation between Log2 LFQ intensities of individual proteins identified in samples purified from RAB3GAP1- and RAB3GAP2-null cells. C, plot to show correlation between Log2 LFQ intensities of individual proteins identified in samples purified from WT and RAB3GAP-null cells. Highlighted datapoints correspond to proteins later found to have RAB3GAP-null:WT intensity ratios ≤0.5. D, Venn diagram to show overlap between all RAB18 associations, TRAPPII-dependent interactions (TRAPPC9-null:WT intensity ratios <0.5), and RAB3GAP-dependent associations (RAB3GAP-null:WT intensity ratios <0.5). E, Western blotting of samples purified from WT and RAB3GAP1-null cells in an independent BioID experiment. Levels of selected proteins are consistent with RAB3GAP-null:WT intensity ratios {braces}.