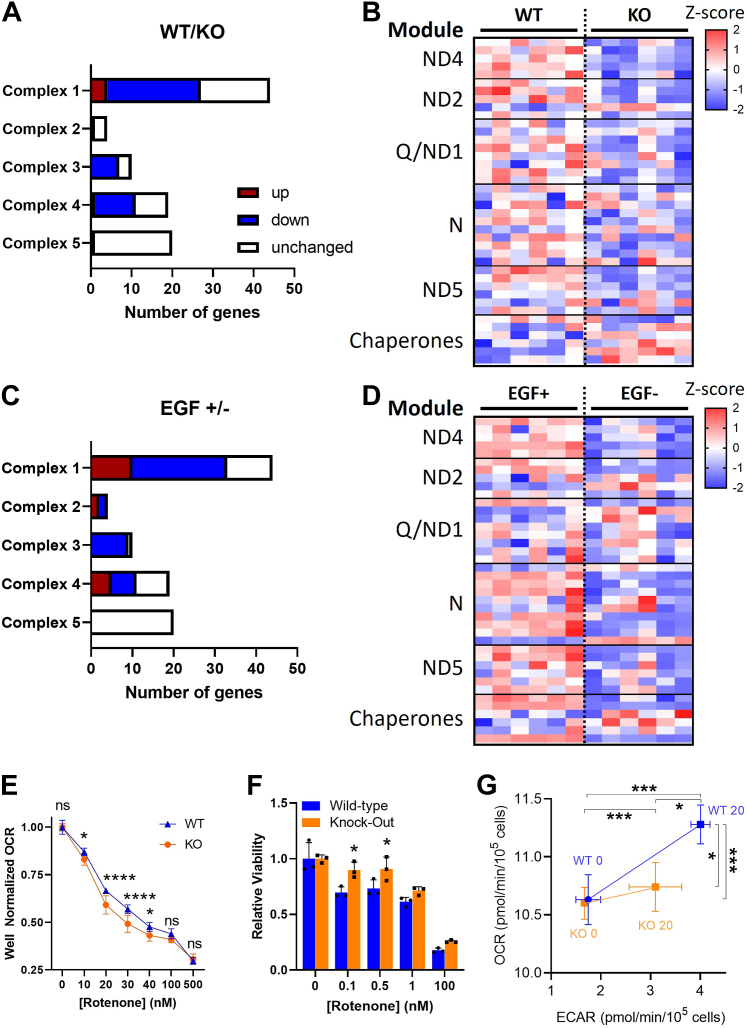
Figure 5.
Mitochondrial complex 1 is transcriptionally and functionally downregulated in PrP KO NSCs.A, mitochondrial gene changes in KO NSCs compared with WT. Statistically increased genes (p < 0.05) are shown in red with decreased genes in blue. B, heatmap displaying Z-scores of complex 1 gene expression in the PrP KO compared with WT NSCs reflecting their assembly modules and select chaperones. C, mitochondrial gene changes in WT NSCs deprived of EGF compared with those in normal levels of EGF. Statistically increased genes (p < 0.05) are shown in red with decreased genes in blue. D, heatmap displaying Z-scores of complex 1 gene expression in WT NSCs deprived of EGF compared with those in normal levels of EGF reflecting their assembly modules and select chaperones. E, Seahorse analysis of oxygen consumption rate (OCR) with increasing concentrations of the complex 1 inhibitor, rotenone, of KO and WT NSCs. F, Prestoblue analysis of cell viability with increasing concentrations of rotenone of PrP KO and WT NSCs. G, energy maps of WT and KO NSCs before and after injection of 1× EGF (20 ng) into the assay media. For B and D, see Supporting informations 3 and 4 for full gene and statistical analyses. For E–G, two-way ANOVA analysis ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, and ∗p < 0.05. Graphs in E and F are displayed relative to the average of the untreated (0 μM/nM rotenone) media conditions for WT or KO NSCs (n = 9 in E and 3 in F). EGF, epidermal growth factor; NSC, neural stem cell; PrP, prion protein.