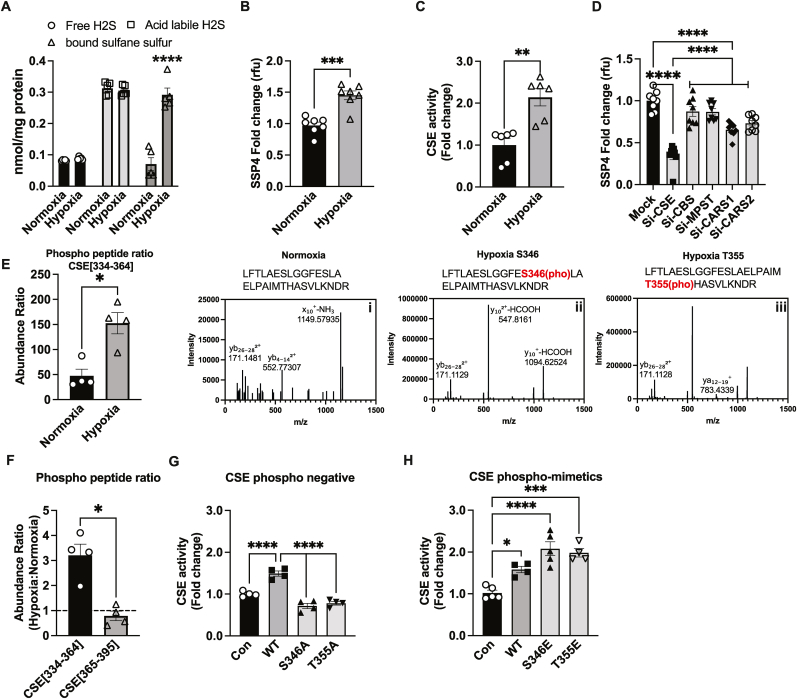
Fig. 1.
Hypoxic per-polysulfide formation and CSE phosphorylation. Panel A shows MAECs exposed to either normoxia (21 % oxygen) or hypoxic (1 % oxygen) conditions for 30min and analyzed for various sulfide metabolites, including free, acid-labile sulfide and bound sulfane sulfur (including persulfide and polysulfide) using MBB/HPLC method. Panel B Per-polysulfides levels in MAECs treated with either normoxia or hypoxia for 30min using the fluorescent probe SSP4. Panel C CSE enzyme activity under hypoxia compared to normoxia in MAECs. Panel D Per-polysulfide levels in MAECs transfected with either mock, CSE, CBS, MPST, CARS-1 or CARS-2 siRNAs and then respectively were probed with SSP4 under hypoxic conditions. Panel E LC/MS HCD fragmentation spectrum of trypsin digested human CSE purified from normoxic versus hypoxic treated HEK cells. CSE amino acid fragment 334–364 (LFTLAESLGGFESLAELPAIMTHASVLKNDR) was identified by Protein Discoverer 2.5. (i) HCD spectra of native Human CSE [334−364] peptide. (ii) HCD spectra of singly phosphorylated Human CSE [334−364] at Ser346. (iii) HCD spectra of singly phosphorylated Human CSE [334−364] at T355. Panel F Phospho peptide ratios comparing CSE[334–364] and CSE[365–395] from LC/MS HCD fragmentation spectrum of trypsin digested human CSE purified from hypoxia. Panel G CSE activity of Control (Con), WT, or CSE phosphonegative alanine mutant constructs of S346A or T355A transfected into HEK293 cells. Panel H CSE activity of HEK293 cells transfected with either Control, WT, CSE glutamate (E) phospho-mimetics, S346E or T355E, under hypoxia. All the data are averaged from triplicates from each experiment with at least n = 5. ****P < 0.0001; ***P < 0.0002; **P < 0.003; *P < 0.01.