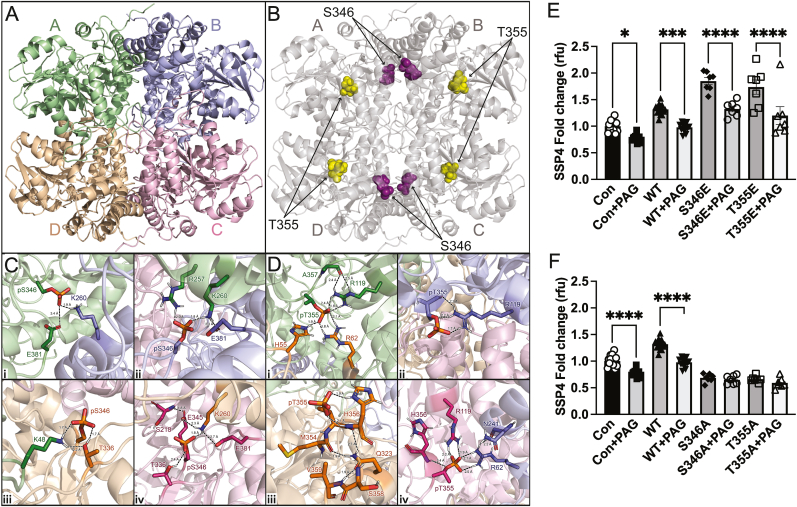
Fig. 5.
Molecular dynamics simulations of cystathionine gamma lyase. Three-dimensional structure of CSE as a tetramer and the respective composite monomers generated by AlphaFold. Molecular dynamics simulations of 350 ns were performed with linear constraint solver (LINCS) constraints for all bonds for panel A WT CSE, and panel B CSE with phospho sites S346 (purple), and T355 (gold). Molecular dynamics simulations Phosphorylation of residues S346 and T355 modeled in Pymol using the PyTMs plugin. All were performed using GROMACS 2019 software with the GROMOS 54A7 force field and SPC216 water model. Frames were recorded every 2 ps. Panel C The 300 ns simulation showing extensive intra- and inter-molecular contacts induced by phosphorylation of 346. Each p346 monomer were color coded belongs to and kept the scheme as shown panel A to illustrate which contacts are intra vs inter-molecular contacts. The backbone RMSD was monitored over the production run of each protein to ensure the stability and convergence of the simulated trajectories. p346 leads to a novel interaction between monomer A (p346) and B (K260, R257), monomer B (p346, H217, E345) and monomer A (K260, R257), monomer C (p346, T336) and monomer A (K48), and monomer D (p346, E345, E381) and monomer C (K260). Panel D Intra- and inter-molecular electrostatic interactions formed by phosphorylation of T355. Interactions to pT355A – H55D, R62D, R119A (Fig. 4D, inset i); pT355B – R119B (Fig. 4D, inset ii); pT355D – Q323D, M354D, H356D, S358D, V359D (Fig. 4d, inset iii); and pT355C – R119C, H356C, R62B, N241B (Fig. 4d, inset iv). Panel E HEK293 cells transfected with either Control (Con), wild type CSE (WT), phospho-mimetic mutants S346E or T355E with or without PAG under hypoxia (1 % oxygen) for 30 min and probed for per-polysulfide (SSP4 fluorophore) Panel F HEK293 cells transfected with either Control (Con), wild type CSE (WT), phospho-negative mutants S346A or T355A with or without PAG under hypoxia (1 % oxygen) for 30 min and probed for per-polysulfide (SSP4 fluorophore). All the data are averaged from triplicates from each experiment with at least n = 5. ****P < 0.0001; ***P < 0.0002; *P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)