Summary
Secondary infection (SI) diagnosis in severe COVID-19 remains challenging. We correlated metagenomic sequencing of plasma microbial cell-free DNA (mcfDNA-Seq) with clinical SI assessment, immune response, and outcomes. We classified 42 COVID-19 inpatients as microbiologically confirmed-SI (Micro-SI, n = 8), clinically diagnosed-SI (Clinical-SI, n = 13, i.e., empiric antimicrobials), or no-clinical-suspicion-for-SI (No-Suspected-SI, n = 21). McfDNA-Seq was successful in 73% of samples. McfDNA detection was higher in Micro-SI (94%) compared to Clinical-SI (57%, p = 0.03), and unexpectedly high in No-Suspected-SI (83%), similar to Micro-SI. We detected culture-concordant mcfDNA species in 81% of Micro-SI samples. McfDNA correlated with LRT 16S rRNA bacterial burden (r = 0.74, p = 0.02), and biomarkers (white blood cell count, IL-6, IL-8, SPD, all p < 0.05). McfDNA levels were predictive of worse 90-day survival (hazard ratio 1.30 [1.02–1.64] for each log10 mcfDNA, p = 0.03). High mcfDNA levels in COVID-19 patients without clinical SI suspicion may suggest SI under-diagnosis. McfDNA-Seq offers a non-invasive diagnostic tool for pathogen identification, with prognostic value on clinical outcomes.
Subject areas: Classification description immunology, Virology, Sequence analysis
Graphical abstract
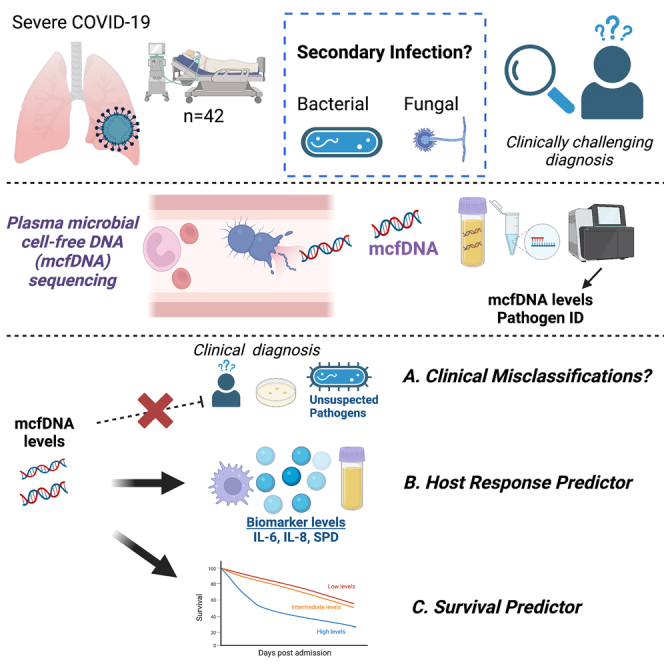
Highlights
-
•
COVID-19 patients exhibit variable levels of plasma microbial cell-free DNA (mcfDNA)
-
•
Plasma mcfDNA levels correlate with inflammation markers and predict poorer survival
-
•
Elevated mcfDNA without clinical infection signs suggests missed secondary infections
-
•
McfDNA sequencing is a promising avenue for pathogen identification and prognostication
Classification Description Immunology; Virology; Sequence analysis
Introduction
Secondary infections complicate the course of up to 50% of hospitalized patients with severe COVID-19 pneumonia and contribute to worse clinical outcomes.1,2,3 Diagnosis of secondary infections (SIs) has presented clinicians with major challenges during the pandemic. Readily available indicators of clinical infection such as fever, leukocytosis, or consolidations on chest imaging cannot distinguish isolated SARS-CoV-2 infection from a secondary pneumonia by super-infecting organisms.4,5 The use of immunomodulators, such as steroids and anti-IL6 receptor inhibitors, has been associated with increased risk for SI, but can also confound clinical and laboratory measurements (e.g., demargination and leukocytosis with steroids or reduction of neutrophils with tocilizumab), further hampering SI diagnosis.6,7,8 Blood cultures are obtained routinely when an SI is suspected in COVID-19, but their diagnostic yield is low, especially in the case of secondary pneumonias, when direct lower respiratory tract (LRT) sampling is recommended.9 LRT sampling for microbiologic studies can be challenging, such as in severely hypoxemic patients on non-invasive respiratory support, and can have low diagnostic sensitivity, due to antecedent antibiotics or slow/fastidious organism growth, as in the case of fungal pathogens.10
Host-response biomarkers have been examined for their prognostic value in COVID-19, as well as their ability to discriminate patients with isolated SARS-CoV-2 infections vs. those with secondary bacterial or fungal infections. Plasma biomarkers in pathways of innate immunity (such as the cytokines interleukin [IL]-6 and IL-8, or the inflammatory molecules CRP and procalcitonin), alveolar epithelial injury (receptor for advanced glycation end products [sRAGE] and Surfactant Protein D [SPD]) or endothelial damage (angiopoietin-2 [Ang-2]) correlate with COVID-19 severity and predict worse outcomes.11,12,13,14,15,16 However, no reliably discriminatory biomarkers for SI have been identified, perhaps due to the complex and heterogeneous host response, the confounding effects of immunomodulator use, the lack of standardized work-up for SIs, and the inherent limitations of conventional microbiologic studies.17
The challenges of SI diagnostic work-up and variable clinical practices have hindered acquisition of accurate SI incidence estimates in severely ill patients with COVID-19. With the prevailing diagnostic uncertainty, empiric antibiotics have been prescribed in 75–85% of hospitalized patients, often initiated due to clinical deterioration and then empirically continued, even in the absence of diagnostic evidence supporting an SI.18,19 Overcoming the diagnostic challenges of SI with reliable, sensitive and non-invasive techniques could optimize diagnostic yield, enabling antibiotic targeting and stewardship.4
Metagenomic sequencing, i.e., the systematic determination of the exact sequence of bases in nucleic acids present in a sample to allow for culture-independent detection of all corresponding microbes, offers novel diagnostic opportunities for infections that are challenging to diagnose. The recent ability to detect and sequence circulating microbial cell-free DNA (mcfDNA-Seq) in plasma has opened the avenue of “liquid biopsy” of infections in sepsis.20 In mcfDNA-Seq of plasma samples, mcfDNA is extracted and sequenced following quantification of human cfDNA (hcfDNA) and filtering out environmental contamination (see STAR Methods for details). The derived mcfDNA sequences are mapped against databases comprising microbial reference genomes which do not contain SARS-CoV-2 sequences, and then quantitative amounts of clinically relevant organisms are reported. Healthy subjects have undetectable (77.2%) or very low mcfDNA levels, whereas patients with clinically suspected sepsis have higher levels of mcfDNA from pathogenic organisms, affording high sensitivity when compared to blood cultures and improved diagnostic yield when compared to composite microbiologic testing methods.20,21 We have previously provided proof-of-concept evidence that patients with COVID-19 with high levels of circulating mcfDNA of common respiratory pathogens were at risk of worse clinical outcome.22 Although the ability to comprehensively assess for SI-causal pathogens with non-invasive blood samples is appealing, feasibility, and clinical validity data are needed. In this study, we report the analysis of a cohort of 42 hospitalized patients with severe COVID-19 who were comprehensively screened for SI pathogens with serial plasma mcfDNA sequencing. We examined the feasibility and clinical validity of mcfDNA-Seq for SI diagnosis, correlation with host response biomarkers, and prediction of outcomes.
Results
Clinical cohort characteristics
We included 42 inpatients with acute hypoxemic respiratory failure due to COVID-19 from April through September 2020 (median age 65.1 years, 62% men, Table 1). We collected plasma samples on enrollment (baseline – day 1) and then repeat samples on days 5 and 10 for patients who remained hospitalized in the Intensive Care Unit (ICU). We obtained a total of 82 plasma samples (median of 2 samples per subject, range 1–5 samples, Figure 1) which we analyzed by mcfDNA-seq, quantification of SARS-CoV-2 RNA levels (vRNA) by qPCR, and measurement of nine host-response biomarkers. Through in-depth review of all available clinical data, we classified patients with regards to presence/absence or clinical suspicion for SI to the following three categories: (1) Microbiologically-Confirmed Secondary Infection (Micro-SI), when typical pathogenic microbes were isolated on clinical biospecimen cultures and antimicrobials were prescribed, (2) Clinically-Diagnosed Secondary Infection (Clinical-SI), when empiric antimicrobials were administered without microbiologic SI confirmation, and (3) “No-Clinical-Suspicion-for-SI” (No-Suspected-SI), when microbiologic workup for SI was negative or not performed, and no empiric antimicrobials were prescribed.
Table 1.
Cohort characteristics for all patients and stratified by secondary infection category
| Variable | All patients | Micro-SI | Clinical-SI | No-Suspected-SI | p-value |
|---|---|---|---|---|---|
| N | 42 | 8 | 13 | 21 | |
| Men, n (%) | 26 (61.9) | 6 (75.0) | 8 (61.5) | 12 (57.1) | 0.68 |
| Age (median [IQR]) | 65.1 [58.4, 76.5] | 61.6 [56.4, 65.6] | 73.5 [61.6, 76.7] | 64.7 [58.1, 76.8] | 0.26 |
| BMI (median [IQR]) | 32.3 [26.5, 39.1] | 32.3 [28.6, 41.1] | 33.5 [30.8, 36.4] | 30.1 [26.2, 38.9] | 0.58 |
| Diabetes, n (%) | 18 (42.9) | 5 (62.5) | 3 (25.0) | 10 (47.6) | 0.17 |
| COPD, n (%) | 10 (23.8) | 2 (25.0) | 3 (25.0) | 5 (23.8) | 0.99 |
| Immunosuppression, n (%) | 4 (9.5) | 1 (12.5) | 0 (0.0) | 3 (14.3) | 0.37 |
| Days from COVID-19 symptom onset (median [IQR]) | 6 [3, 9] | 6 [3, 9] | 7 [5, 9] | 5 [1, 9] | 0.53 |
| Glucocorticoid Use, n (%) | 24 (57) | 4 (50) | 9 (70) | 11 (52) | 0.65 |
| Invasive Mechanical Ventilation, n (%) | 27 (64.3) | 7 (87.5) | 5 (38.5) | 15 (71.4) | 0.05 |
| ECMO, n (%) | 8 (19.5) | 3 (37.5) | 1 (7.7) | 4 (19.0) | 0.24 |
| WBC (median [IQR]) | 10.2 [6.7, 12.4] | 13.3 [10.0, 14.2] | 6.7 [5.9, 11.8] | 10.6 [7.4, 11.7] | 0.04 |
| WHO ordinal scale at ICU admission (median [IQR]) | 8.0 [7.2, 9.0] | 7.0 [5.0, 9.0] | 7.0 [6.0, 7.0] | 6.0 [5.0, 6.0] | 0.22 |
| RALE score (median [IQR]) | 24.0 [21.1, 31.2] | 21.8 [20.6, 23.1] | 22.2 [21.0, 32.6] | 26.2 [23.0, 35.8] | 0.10 |
| SARS-CoV-2 RNA in Plasma (cps/mL, median [IQR]) | 867.0 [40.0, 5,742.5] | 3.0 [3.0, 3.0] | 1,795.5 [86.5, 6,840.0] | 2,491.0 [220.0, 5,445.0] | 0.05 |
| SARS-CoV-2 RNA in ETA (cps/mL, median [IQR]) | 215,466.7 [2,718.8, 729,422.2] | 29.0 [29.0, 2,804.8] | 338,240.0 [17,920.0, 508,444.4] | 339,911.1 [33,351.1, 950,400.0] | 0.18 |
| IL-6 pg/mL (median [IQR]) | 37.1 [15.7, 159.5] | 128.0 [66.9, 188.4] | 26.9 [14.7, 41.4] | 36.0 [16.8, 165.7] | 0.44 |
| IL-8 pg/mL (median [IQR]) | 22.8 [14.7, 41.4] | 29.1 [19.9, 52.4] | 13.9 [7.1, 20.0] | 29.9 [17.6, 53.6] | 0.05 |
| ST2 pg/ml (median [IQR]) | 190,532.0 [95,113.3, 268,265.7] | 190,174.0 [89,313.7, 276,629.2] | 206,023.5 [151,105.8, 270,770.7] | 185,961.2 [97,840.6, 253,415.2] | 0.79 |
| TNFR1 pg/ml (median [IQR]) | 4,539.0 [3,500.3, 8,877.2] | 7,622.3 [4,678.7, 11,256.5] | 4,072.4 [3,179.9, 5,512.4] | 4,498.0 [3,390.3, 9,493.9] | 0.28 |
| SPD pg/ml (median [IQR]) | 25.1 [14.0, 57.6] | 40.0 [21.9, 83.8] | 39.0 [27.0, 55.1] | 20.5 [8.3, 29.7] | 0.26 |
| RAGE pg/ml (median [IQR]) | 4,576.8 [2,422.6, 9,071.2] | 2,519.2 [1,566.5, 8,520.3] | 2,689.6 [2,383.5, 8,455.6] | 5,879.8 [3,856.5, 9,982.9] | 0.17 |
| Ang-2 pg/mL (median [IQR]) | 4,354.0 [2,484.3, 7,375.3] | 9,805.2 [5,706.4, 14,848.5] | 3,292.3 [2,098.3, 7,299.0] | 3,195.6 [2,422.8, 5,096.5] | 0.01 |
| Procalcitonin pg/ml (median [IQR]) | 4,61.0 [187.9, 1,516.1] | 403.8 [164.1, 798.8] | 206.1 [147.6, 454.8] | 815.1 [272.2, 1,767.8] | 0.08 |
| Pentraxin-3 pg/mL (median [IQR]) | 7,826.2 [3,584.4, 16,080.0] | 6,908.7 [2,682.9, 8,793.6] | 6,827.5 [3,649.9, 11,186.3] | 13,548.5 [3,976.4, 20,466.9] | 0.14 |
| Human cfDNA MPM (median [IQR]) | 563.5 [148.6, 1,284.8] | 750.6 [200.8, 1,299.4] | 325.9 [109.1, 563.7] | 686.2 [148.7, 2,253.5] | 0.17 |
| Total microbial cfDNA MPM (median [IQR]) | 812.9 [151.3, 2,435.5] | 1,614.9 [794.8, 1,7812.8] | 1.0 [1.0, 818.1] | 1,026.6 [457.7, 2,464.9] | 0.17 |
| Bacterial cfDNA MPM (median [IQR]) | 600.4 [1.0, 2,906.1] | 9,349.2 [559.7, 28,503.5] | 1.0 [1.0, 86.0] | 822.9 [117.9, 1,956.0] | 0.07 |
| Fungal cfDNA MPM (median [IQR]) | 1.0 [1.0, 1.0] | 158.8 [1.0, 388.7] | 1.0 [1.0, 1.0] | 1.0 [1.0, 1.0] | <0.01 |
| Respiratory pathogen cfDNA MPM (median [IQR]) | 1.0 [1.0, 1,480.8] | 9,349.2 [559.7, 28,503.5] | 1.0 [1.0, 13.5] | 118.0 [1.0, 1,480.8] | 0.06 |
Abbreviations: Microbiologically Diagnosed Secondary Infection (Micro-SI); Clinically Diagnosed Secondary Infections (Clinical-SI); No Clinical Suspicion for SI (No-Suspected-SI); chronic obstructive pulmonary disease (COPD); extracorporeal membrane oxygenation (ECMO); Radiographic Assessment of Lung Edema score (RALE score); receptor for advanced glycation end products (RAGE); suppression of tumorigenicity (ST-2); tumor necrosis factor receptor (TNFR-1); Surfactant Protein D (SPD); molecules per microliter (MPMs).
p values significant below threshold of 0.05 are shown in bold. See also Figures S1–S6.
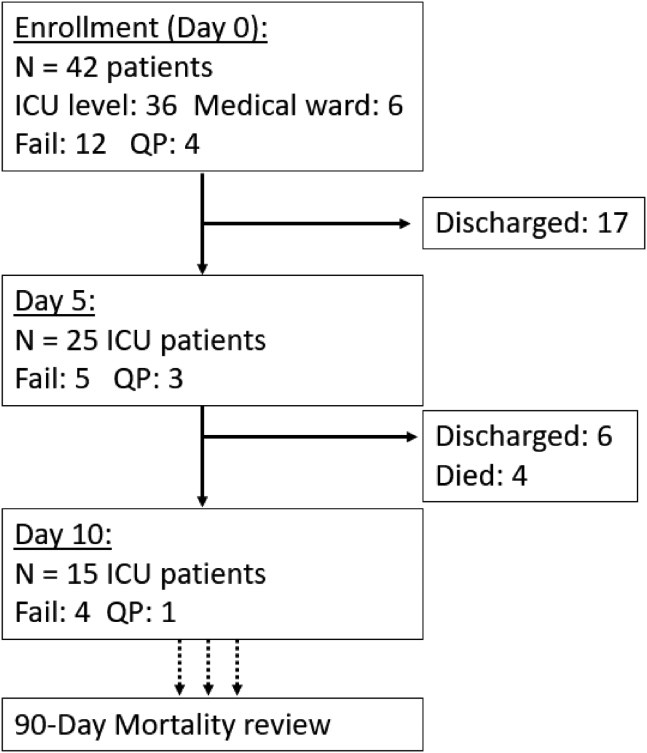
Figure 1.
Flowchart of enrolled patients sampled at each follow-up period through the 90-day mortality endpoint
Fail: failed mcfDNA sequencing; QP: qualitative pass.
Of the 42 patients included, eight patients (19.5%) were supported on extracorporeal membrane oxygenation (ECMO), and 27 (64.3%) were invasively mechanically ventilated (IMV, Table 1). At baseline, we classified subjects as Micro-SI (n = 8), Clinical-SI (n = 13), and No-Suspected-SI (n = 21) (Table 1). We found lower rates of IMV (p = 0.05) and lower total white blood cell count (p = 0.04) in Clinical-SI patients, but no other significant differences in clinical variables, including ECMO status, between SI groups. Plasma SARS-CoV-2 vRNA was notably lower in Micro-SI (p ≤ 0.05), although there was no difference in time (days) from COVID-19 symptom onset to biospecimen acquisition between clinical groups (Table 1). In biomarker comparisons, we found lower IL-8 and procalcitonin levels in patients with No-Suspected SI vs. Clinical-SI, and overall higher levels of Ang-2 in Micro-SI patients (Table 1, Figure S1). Similarly, IL-8 and procalcitonin differed significantly by clinical severity on admission based on the WHO ordinal scale (Figure S2), whereas comparisons by the level of respiratory support required at time of sampling (ECMO, IMV, or non-invasive) showed that patients on ECMO has significantly higher levels of IL-6 and Ang-2 compared to patients on non-invasive respiratory support (Figure S3).
Output of plasma metagenomic sequencing runs
We conducted plasma mcfDNA-Seq with the Karius Test® [Karius Inc., Redwood City, CA] and classified the derived metagenomic sequences as human (hcfDNA) vs. microbial (mcfDNA). Based on minimum sequencing coverage metrics required for quality control (see STAR Methods for details), we classified sequencing runs for the 82 samples across the study period “Pass” (i.e., successful, n = 52, 63%), “Qualitatively Pass” (n = 8, 10%) or “Failed” (n = 22, 27%, eTable 1). Notably, “Pass” samples had significantly lower levels of hcfDNA compared to “Qualitative Pass”, or “Fail” samples (p < 0.0001, Figure S4), and these differences in hcfDNA levels by sequencing run success were independent of whether samples were taken from patients on ECMO or not (Figure S5). We also found that a successful sequencing run (“Pass”) on a day 1 sample for a given subject was significantly associated with a “Pass” run on a day 5 sample (Fisher’s odds ratio 11.9, 95% confidence interval-CI [1.00–703.8], p = 0.04). From “Qualitative Pass” samples, we utilized only qualitative information about identified microbial species, whereas we utilized quantitative mcfDNA data (expressed as molecules per microliter, MPMs) from “Pass” samples only.
Plasma metagenomics results by SI category
Among the 52 “Pass” samples across all time-points, Micro-SI diagnosis had more samples positive for mcfDNA (i.e., mcfDNA sequencing reads that mapped to at least one microbial species reported, 15/16 [94%] samples) compared to Clinical-SI (7/13 [57%], p = 0.02, Table S2). Unexpectedly, a high proportion of No-Suspected-SI samples, including “Pass” and “Qualitative Pass”, were positive for mcfDNA (25/30 [83%]). All eight “Qualitative Pass” samples (seven No-Suspected-SI samples and one Clinical-SI sample), were positive for mcfDNA, with 3/8 samples reporting ≥2 microbial species.
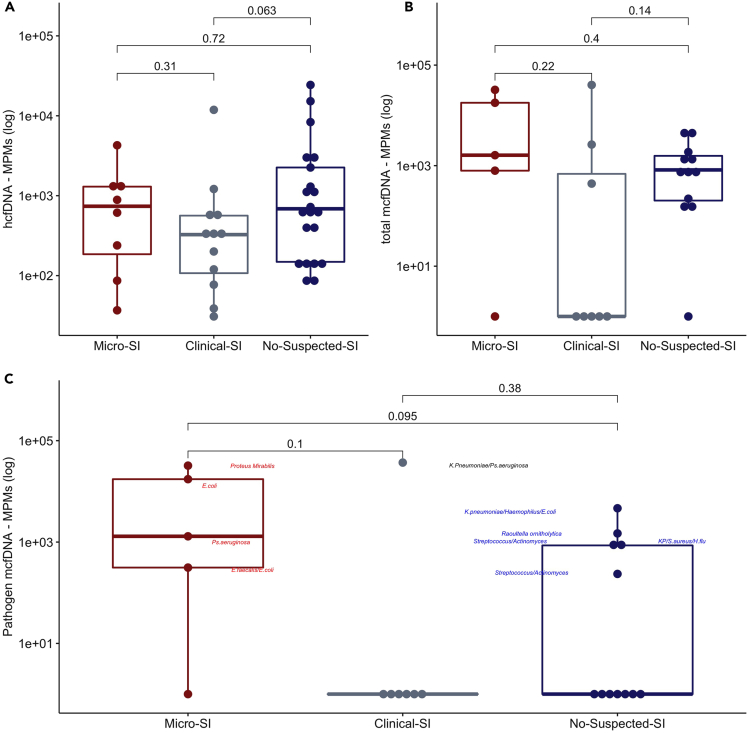
Baseline samples were collected at a median (interquartile range – IQR) of 3 (3) days from hospital admission. Of the 25 baseline “Pass” samples, 72% were positive for mcfDNA, with a median (IQR) of 812.9 (151.3–2,435.5) MPMs per sample, 59.1% of which corresponded to typical pathogenic organisms. Stratified by SI categories, 4/5 (80.0%) Micro-SI, 3/8 (37.5%) Clinical-SI, and 11/12 (91.6%) No-Suspected-SI samples were positive for mcfDNA, with a statistically significant higher proportion of positive samples in No-Suspected-SI compared to Clinical-SI samples (Fisher’s p = 0.01). We found no significant differences in baseline hcfDNA, total mcfDNA and pathogen mcfDNA MPMs between SI groups (Figures 2A–2C), although Micro-SI samples had numerically higher total and pathogen mcfDNA MPM levels (Table 1).
Figure 2.
Clinical classification of secondary infection diagnosis among patients with COVID-19 and plasma cell-free DNA levels
Clinical classification of secondary infection diagnosis among patients with COVID-19 did not show significant differences in baseline human (A) or microbial cell-free DNA levels (B and C) (N = 42). Subjects classified as microbiologically confirmed secondary infection (Micro-SI) had numerically higher but statistically non-significant different baseline levels of total and pathogen microbial cell-free DNA (mcfDNA) compared to subjects classified as clinically diagnosed secondary infections (Clinical-SI) or those with no clinical suspicion for SI (No-Suspected-SI). Pairwise comparisons were conducted with Wilcoxon test. In C, the combined pathogen mcfDNA for each sample is shown, with the most abundant microbial species in each of the positive samples denoted beside the sample data points. See also Figures S4–S6, and Table S2. Data in boxplots are represented as individual values with median values and interquartile range depicted by the boxplots.
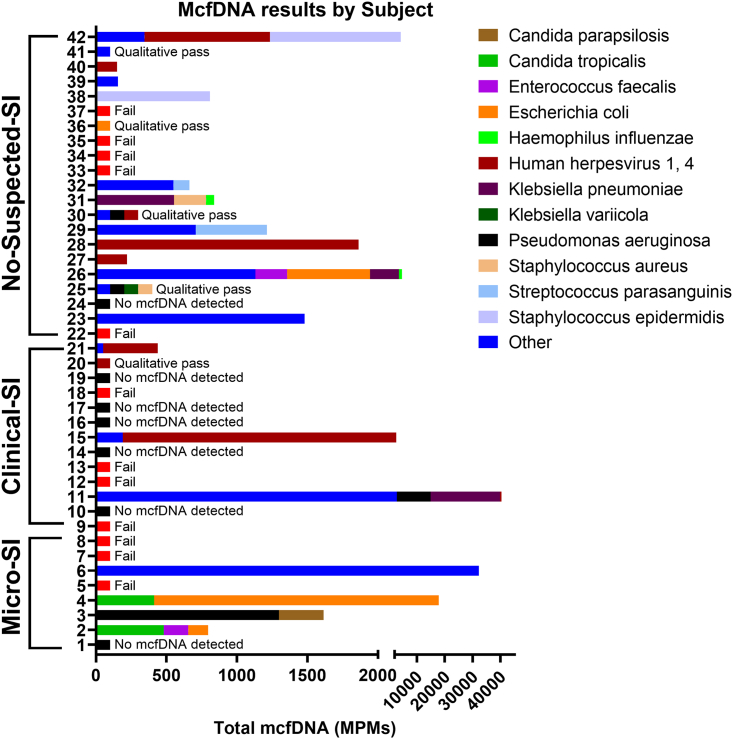
Among baseline “Pass” samples in Micro-SI subjects, plasma metagenomics reported mcfDNA from the culprit organisms identified by cultures in 3/5 (60.0%) cases (Escherichia coli in subject 4, Pseudomonas aeruginosa in subject 3, and Proteus mirabilis in subject 6) (Figure 3, Table S3). The two discordant Micro-SI samples involved two probable ventilator-associated pneumonia (VAP) cases: subject 1 with negative mcfDNA-Seq when LRT cultures taken three days prior to plasma sampling showed rare E. coli growth, and subject 2 with light Methicillin-sensitive Staphylococcus aureus growth in LRT cultures taken two days prior to plasma sampling was deemed the culprit, but mcfDNA-Seq reported Enterococcus faecalis and E. coli mcfDNA. When considering all available Micro-SI samples (five baseline and 11 follow-up samples), mcfDNA-Seq results were concordant with cultures in 13/16 (81.3%) of comparisons.
Figure 3.
Significantly higher proportion of positive samples with mcfDNA species calls in No-Suspected-SI compared to Clinical-SI samples.
We display samples grouped by SI classification and sample identifier, showing the species and respective MPMs called by mcfDNA-Seq. Standardized bars indicate failed samples, samples with no species called, and those with qualitative pass, yielding species each denoted with standardized bar. Infrequently called species were combined for visual simplicity.
Baseline samples from Clinical-SI subjects contained mcfDNA in 37.5% of cases (3/8). Subject 11 had high mcfDNA levels for plausible pathogens (Klebsiella pneumoniae and P. aeruginosa; Figure 3, and Table S3), which had not been detected by blood cultures (no LRT specimen cultures available). For the other two Clinical-SI subjects (15 and 21), reported mcfDNA belonged mostly to Human Herperviruses, which could represent viral re-activation in context of critical illness.23
Among No-Suspected-SI patients at baseline, 11/12 (91.6%) were positive for mcfDNA, with five samples showing >100 pathogen mcfDNA MPMs in the range of Micro-SI subjects (annotated species in Figures 2C and 3). The remaining six subjects with mcfDNA revealed organisms of unclear clinical significance: Herpesvirus DNA in four subjects (27, 28, 38, and 42), and gram-positive bacteria often considered as skin contaminants in blood cultures (Lactobacillus gasseri and Staphylococcus epidermidis) in three subjects (38, 41, and 42). Notably, 3/4 (75%) of the baseline “Qualitative Pass” samples for No-Suspected-SI subjects (25, 30, 36) were also positive for pathogen mcfDNA, even though the absolute burden of mcfDNA MPMs could not be reliably estimated.
Comparison of circulating cfDNA burden in COVID-19 vs. historic non-COVID samples
Comparing against published data from our group for mechanically ventilated patients with microbiologically confirmed pneumonia (n = 26), clinically diagnosed pneumonia (n = 41) and uninfected controls (n = 16, intubated for airway protection or due to cardiogenic pulmonary edema), we found markedly higher levels of hcfDNA in subjects with COVID-19 compared to all non-COVID-19 patient groups (COVID-19 median 563.5 MPMs [148.6, 1,284.8]; non-COVID-19 median 120.9 MPMs [56.5, 529.4], Figure S6A, p < 0.005).24 All non-COVID patients were supported by IMV but none were on ECMO. Non-COVID patients with microbiologically confirmed pneumonia had higher total and pathogen mcfDNA levels compared to No-Suspected-SI COVID-19 patients (total mcfDNA median 4,085 MPMs [1,553, 39,970.2] vs. median 1,026.6 MPMs [457.7, 2,464.9], respectively p = 0.015), who in turn had markedly higher total and pathogen mcfDNA levels compared to uninfected controls (total mcfDNA median 0 MPMs [0, 48.0], p = 0.00018, Figures S6B and S6C).
Trajectories of plasma cfDNA levels in subjects with COVID-19
We reclassified subjects by SI status at the time of the follow-up sample acquisition and found that few subjects transitioned into a different SI diagnostic category (4/26 [15%] and 4/16 [25%] for days 5 and 10, respectively, Figures S5 and S7). At post-enrollment day 5, Micro-SI subjects had significantly higher total and pathogen mcfDNA levels compared to No-Suspected-SI subjects (p < 0.05, Figure S7).
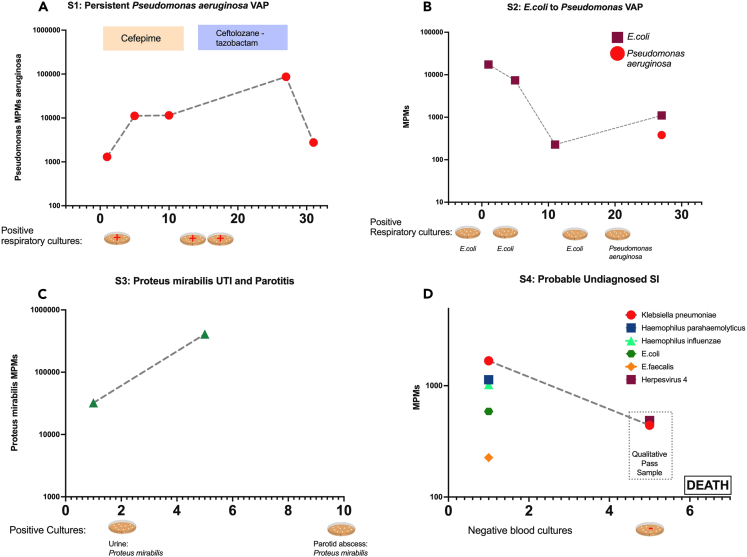
We examined subject-level trajectories in the four cases with persistent culture-proven SI or persistent pathogen mcfDNA detection on available longitudinal samples (Figure 4). Subject 3 was diagnosed with resistant Pseudomonas aeruginosa VAP with clinical and microbiologic relapse, demonstrating persistently elevated Pseudomonas aeruginosa mcfDNA for >30 days on repeated sampling (Figure 4). For subject 4 McfDNA-Seq demonstrated a transition in detected bacterial species in follow-up samples corresponding to two different VAP episodes (by E. coli and Pseudomonas aeruginosa respectively). Subject 6 had unexplained, persistent Proteus mirabilis mcfDNA detection following treatment of a urinary tract infection, but was found to have a late parotid gland abscess that was drained and cultures grew Proteus mirabilis. Last, for subject 26 diagnosed as No-Suspected-SI (i.e., no empiric antimicrobials) who died on day 6 with multi-organ failure attributed to isolated SARS-CoV-2 infection, we found high pathogen mcfDNA levels, including Klebsiella pneumoniae, raising concern for the possibility of an undetected and undertreated SI.
Figure 4.
Comparison of plasma mcfDNA sequencing with microbiologic/clinical diagnoses across the timeline of four subjects with serial sampling
Species-specific microbial MPMs (y axis) are shown across sequential sampling days from enrollment, (x axis). Petri dish graphics along the x axis denote the timing and results of clinically obtained microbiological testing. Subject 3 (A) was an immunocompetent patient with a persistent culture-confirmed Pseudomonas VAP that persisted through the antimicrobials, with timing and therapy noted above. Subject 3 had persistently elevated Pseudomonas mcfDNA levels throughout the extended infection course, which ultimately correlated with clinical improvement (A). Subject 4 (B) had sequential VAPs with changing pathogens detected on invasive respiratory cultures, E. coli followed by Pseudomonas, which was concordantly reported on noninvasive mcfDNA sampling. Subject 6 (C) was a patient with diabetes who was initially found to have a resistant Proteus urinary infection, and a subsequent persistent septic clinical picture, later found to have a polymicrobial, including Proteus, parotid gland abscess. Proteus mcfDNA levels remained elevated in subject 6 during initial antimicrobial therapy for urinary infection, suggesting the persistent source of infection. Subject 26 (D) was clinically determined No-Suspicion-for-SI, but had a deteriorating clinical course, without cultures obtained for 5 days or empiric antimicrobials, and ultimately died of shock and multisystem organ failure. Noninvasive testing revealed persistent levels of Klebsiella pneumonia mcfDNA levels, which suggests an undiagnosed SI may have contributed to the clinical course. See also Figure S7.
McfDNA is associated with plasma host-response biomarkers and LRT bacterial burden
We next examined the relationship between mcfDNA-Seq results and clinical/laboratory parameters of COVID-19 severity. We found no significant correlation between mcfDNA and SARS-CoV-2 viral load (quantified by vRNA qPCR in plasma and endotracheal aspirate samples, when available), radiographic severity (quantified by radiographic assessment of lung edema [RALE] scores) or clinical severity by the WHO ordinal scale.25,26,27 However, there was a weak, significant correlation between hcfDNA and RALE scores (r = 0.33, p = 0.03).
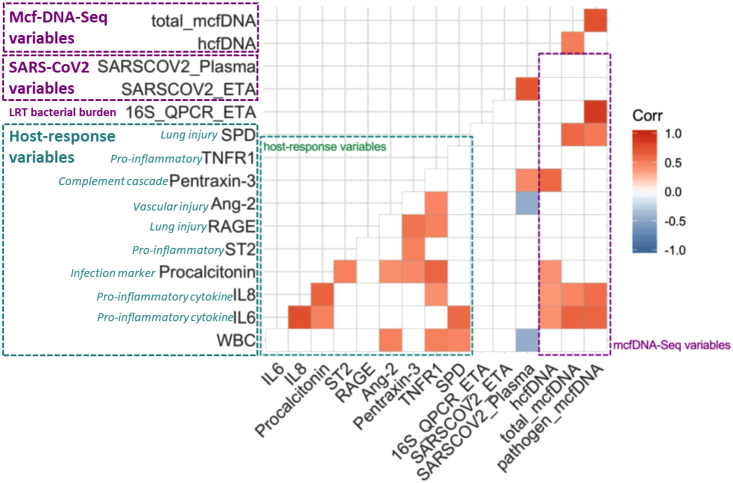
We then examined for correlations between cfDNA levels and baseline plasma biomarkers of host response. Total and pathogen mcfDNA levels were significantly correlated with the cytokines IL-6 and IL-8, as well as SPD levels, the innate host-defense glycoprotein secreted by alveolar cells. The associations for IL-6 and SPD with mcfDNA remained significant even following adjustment for vRNA levels (Table S4), suggesting effects of mcfDNA on host response independent of SARS-CoV-2. HcfDNA levels were significantly correlated with IL-6, IL-8, pentraxin-3, and procalcitonin levels following adjustment for multiple comparisons (Figure 5). Pathogen mcfDNA levels were significantly correlated with bacterial 16S rRNA gene copies by qPCR in available endotracheal aspirate specimens (n = 19), a surrogate of LRT bacterial load (Pearson’s r = 0.92, p = 0.0004).
Figure 5.
Baseline plasma microbial cell-free DNA levels were significantly correlated with lower respiratory tract bacterial load and plasma host response biomarkers (N = 42).
Correlogram demonstrating comparisons of host nine response biomarkers (green dashed box), number of 16S rRNA gene copies by qPCR in Endotracheal Aspirates (ETA, surrogate for lower respiratory tract bacterial load), number of SARS-COV-2 RNA copies in ETA and plasma samples by qPCR, and mcfDNA-Seq output (hcfDNA, total mcfDNA and pathogen mcfDNA – purple dashed box). Significant correlations with the Pearson’s method and following adjustment for multiple comparisons by the Benjamini-Hochberg method are shown, with direction and strength of correlation depicted by the color scale on the right panel. See also Table S4.
Baseline mcfDNA levels are predictive of 90-day survival
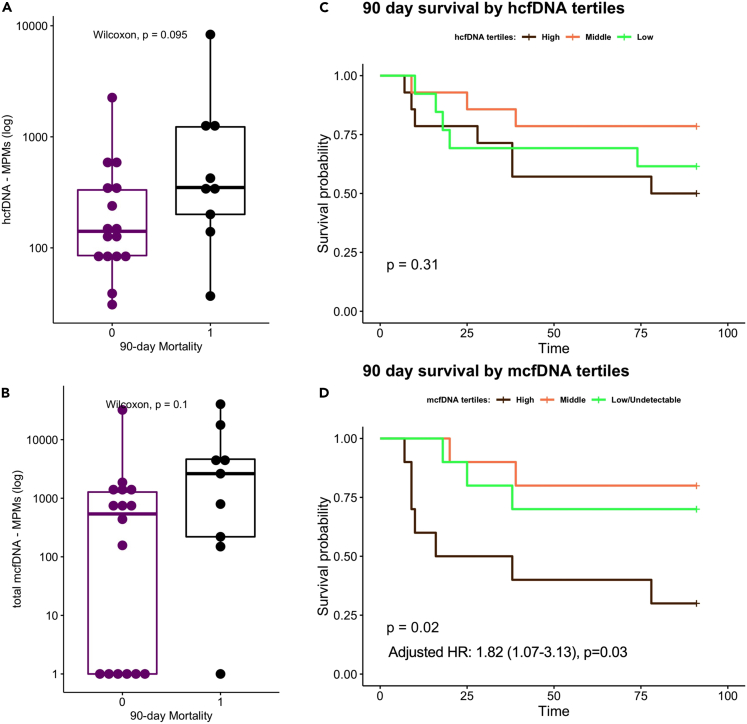
Baseline hcfDNA and total mcfDNA levels were not significantly associated with cumulative mortality at 90 days from ICU admission (Figures 6A and 6B). Baseline total mcfDNA levels were significantly associated with worse 90-day survival in a Cox proportional hazards model adjusted for age (hazard ratio for log10-transformed mcfDNA 1.82, [1.07–3.13], p = 0.03). Stratified by baseline total mcfDNA tertiles for visualization of survival curves (high>1,499, middle 344–1,499, low<344 MPMs), subjects in the high mcfDNA tertile had worse 90-day survival by Kaplan-Meier curve analysis compared to subjects in other tertiles (global log rank p = 0.02), whereas hcfDNA tertiles were not predictive of 90-day survival (Figures 6C and 6D). In contrast to the significant prognostic information of baseline total mcfDNA levels, there were no 90-day survival differences between the clinical classification SI groups (log rank p = 0.62 for Micro-SI, Clinical-SI, and No-Suspected-SI, data not shown). In a Cox model adjusted for age and SI classification, total mcfDNA levels remained significantly associated with worse 90-day survival (hazard ratio for log10-transformed mcfDNA 2.23, [1.25–3.98], p = 0.006), with no significant effect modification by SI group (non-significant interaction terms in the Cox model). Pathogen mcfDNA levels showed prognostic associations in similar direction with total mcfDNA levels, although results were not statistically significant (hazard ratio for log10-transformed pathogen mcfDNA 1.31, [0.96–1.78], p = 0.08). Stratified by 90-day survival, we found no significant differences in the longitudinal trajectories of total mcfDNA and hcfDNA levels between survivors and non-survivors (Figures S8A and S8B).
Figure 6.
Highest microbial cell-free DNA levels at baseline were significantly associated with worse 90-day survival (N = 42).
90-day non-survivors had numerically higher (but statistically non-significant) plasma hcfDNA (A) and mcfDNA (C) levels. Data in boxplots are represented as individual values with median values and interquartile range depicted by the boxplots. HcfDNA levels were not significantly associated with 90-day survival by Kaplan-Meier analysis (B). Patients with the highest tertile of mcfDNA (>1499 Molecules per Microliter) had significantly worse survival compared to patients in the other two mcfDNA tertiles (D). In a Cox proportional hazards model adjusted for age, mcfDNA levels were significantly associated with increased hazards of death (D, adjusted hazard ratio for log10-transformed mcfDNA 1.82, 95% confidence interval 1.07-3.13, p = 0.03). See also Figure S8.
Discussion
In this translational study, we systematically evaluated 42 COVID-19 inpatients with culture-independent mcfDNA-Seq, viral load quantification and host response profiling. We found no significant differences in mcfDNA levels by SI clinical categories at baseline, although culture-confirmed cases of SI (Micro-SI) had higher mcfDNA levels on follow-up samples. The mcfDNA species corresponded to clinically isolated pathogens in 81% of Micro-SI samples, and indicated potentially missed, super-infecting pathogens in up to 41% of subjects for whom there was no clinical suspicion for SI (No-Suspected-SI). Our molecular analyses revealed significant correlations of circulating mcfDNA levels with LRT bacterial burden, suggestive of plausible secondary pneumonias, as well as with plasma host-response biomarkers. Patients with higher mcfDNA had significantly lower 90-day survival, independent of SI group. Longitudinal assessments revealed that mcfDNA levels can persist in SI without adequate source control, such as the patient with parotitis, or with antibiotic resistant pathogens, such as the patient with resistant Pseudomonas VAP. Thus, serial mcfDNA-Seq may offer a non-invasive tool for monitoring treatment response, although further research is needed to understand the temporal kinetics of mcfDNA levels in infections that respond to treatment.
Our results were notable for an unexpectedly high rate of failed mcfDNA-Seq runs due to high burden of circulating hcfDNA, which was much higher than prior cases of non-COVID-19 pneumonia. The high failure rate in this cohort of COVID-19 subjects cannot be explained by technical or processing issues because we used the same protocols and processes as in prior and ongoing studies (GDK, unpublished data) with mcfDNA-Seq with sequencing success rates of more than 90%. The high amounts of interfering hcfDNA provide the most plausible explanation for the 27% sequencing failure rate in this study. Such high hcfDNA levels likely reflect substantial human tissue damage in severely ill patients with COVID-19, a finding consistent with another study showing higher hcfDNA in COVID-19 compared to patients with influenza or respiratory syncytial virus infections, despite similar clinical severity indices.24,28 In our cohort, samples from patients on ECMO, who have more severe lung injury and at the same time their circulating WBCs are continuously exposed to the sheer stress of extra-corporeal circulation that may result in lysis and release of hcfDNA, had numerically higher sequencing failure rates, although hcfDNA levels were not significantly different from patients not on ECMO. The higher likelihood of subsequent failed samples after initial failed samples suggests subject-specific influences on sequencing run outputs and results.
Our study highlights the challenge of clinically distinguishing cases with isolated SARS-CoV-2 infection from those complicated by SIs. The results of host-response biomarker analyses and mcfDNA-Seq did not align with the clinical SI diagnoses or the microbiologic investigations guided by clinical judgment. For example, although procalcitonin levels are commonly used as diagnostic biomarkers for bacterial infections, their diagnostic value for SIs in COVID-19 remains uncertain.14,29 Notably, we observed higher procalcitonin levels in No-Suspected SI compared to Clinical-SI patients, which suggests a potential misclassification of cases by clinicians. However, we found no significant correlation between procalcitonin and mcfDNA levels. Consequently, procalcitonin exhibited limited validity in our dataset for distinguishing between SI and isolated SARS-CoV-2 infection. The clinical validity of mcfDNA sequencing was further substantiated by its significant correlations with LRT bacterial burden and systemic host-response biomarkers. Additionally, mcfDNA sequencing demonstrated predictive value in determining 90-day survival. Our study suggests that noninvasive mcfDNA sequencing may provide valuable insights into the presence of SIs and their impact on patient outcomes.
Circulating mcfDNA was significantly associated with host immune response in COVID-19. McfDNA species may reflect SI-causal pathogens (regardless of their viability in the bloodstream), but may also signify lung barrier disruption in patients with acute lung injury from COVID-19, or even gut barrier disruption in patients with circulatory shock and hypoperfusion.24 Thus, positive mcfDNA calls by the Karius test should not be directly interpreted as evidence of superinfecting pathogens, but interpreted within the context of critical illness with colonized mucosal surfaces by microbiota and impaired barrier function. The significant correlation between hcfDNA levels and RALE scores suggests that lung injury may account, at least in part, for the hcfDNA levels, although we could not experimentally evaluate the tissue origin of circulating hcfDNA. Additionally, quantitative assessments of hcfDNA may have been influenced by the host DNA depletion steps during sample processing and library preparation for mcfDNA-Seq. Recent research has highlighted the relevance of hcfDNA as a diagnostic/prognostic biomarker in sepsis, with higher levels of deoxyribonuclease 1 (DNase 1) and cfDNA in preterm infants with early onset neonatal sepsis compared to controls, as well as altered hcfDNA fragmentation patterns in adult patients with sepsis, perhaps due to diverse tissue origin and multiorgan damage.30,31 Therefore, further study of the human subset of the circulating cfDNA may offer further insights in critical illness diagnosis and prognostication.
On the microbial side, the significant correlations of mcfDNA with IL-6 and IL-8 levels highlighted the potential importance of mcfDNA in the inflammatory cascade of COVID-19, since mcfDNA can be recognized by Toll-like receptors and propagate systemic inflammatory responses, independent of the effects of SARS-CoV-2 on innate immunity.32 SPD, a critical lung innate immunity mediator involved in binding of microbes for phagocyte recognition and clearance, was also significantly correlated with mcfDNA. SPD has shown predictive value in COVID-19, and we have recently shown that lung microbiota clusters are significantly associated with plasma SPD levels in patients with non-COVID pneumonia.33,34 Our data now connect a circulating microbial biomarker (mcfDNA) with plasma levels of a lung-origin molecule (SPD), suggesting that non-invasive study of host-microbiota interactions in the plasma can offer insights into LRT processes. The case of Herpesvirus DNA identification highlighted the prevalence of viral re-activation in critical illness. Consequently, despite the prognostic impact of measured mcfDNA levels in patients with COVID-19, the use of mcfDNA-Seq as diagnostic tool for SIs requires careful interpretation. The detected mcfDNA signal may represent superinfecting bacterial or fungal VAP pathogens, translocating commensal microbes, sample collection skin/environmental contaminants or re-activated viral organisms, and distinction between such possibilities necessitates an integrative assessment by clinicians.
Our study advances understanding of the biologic and diagnostic importance of circulating mcfDNA in COVID-19 and demonstrates limitations in clinical assessment of SIs. McfDNA correlated with biomarkers of host immune response and LRT microbial burden. The noninvasive modality of quantifying mcfDNA load and accurately identifying microbial species may offer a sensitive tool for SI detection, as well as further advance our understanding of the role of translocating microbiota in critical illness. McfDNA sequencing may be particularly helpful in spontaneously breathing patients on high flow nasal cannula or non-invasive mechanical ventilation, for whom access to LRT specimens is challenging. Further prospective investigation for treatment guidance is necessary to systematically evaluate the incidence of molecular evidence for SI and examine the impact of non-invasive screening on patient outcomes.
Limitations of the study
Our study has several limitations. The sample size of 42 subjects of our exploratory analysis limits the statistical power for some of the analyses, yet it is to our knowledge the largest study to utilize plasma metagenomics in patients with COVID-19, and we found significant associations with host inflammation and outcomes, consistent with our hypotheses.35 Clinical samples for microbiologic workup were collected at the discretion of the treating clinicians, and thus some of the mcfDNA-culture comparisons are limited. Nonetheless, our clinical dataset is representative of standard of care at a tertiary academic center. We systematically examined all patients for molecular evidence of possible SI using systematic evaluation with mcfDNA testing, regardless of clinician impressions. It is possible that for certain No-Suspected-SI subjects, the mcfDNA reported may not represent an active infection. Careful review of the mcfDNA-Seq output and integration with available evidence on organismal pathogenicity can help further inform interpretation of pathogen mcfDNA. Nonetheless, circulating mcfDNA carries prognostic information underlined by the significant associations with systemic inflammation and survival.22 Our study was limited by an unexpected amount of failed mcfDNA-Seq analyses due to high amounts of interfering hcfDNA, but this finding provided important insight into the high degree of human cellular damage in COVID-19, and inclusion of subjects with failed mcfDNA-Seq sequencing offers full disclosure of our cohort study and the range of expected analyzable results from future clinical implementation of mcfDNA-Seq testing. Finally, our cohort was enrolled early in the pandemic, prior to wide use of steroids and additional immunomodulators for COVID-19, which have been associated with higher rates of SI and could further alter the measured host immune responses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Endotracheal aspirates, Plasma samples | University of Pittsburgh, Division of Pulmonary, Allergy, Critical Care And Sleep Medicine |
Details provided in this manuscript |
| Critical commercial assays | ||
| ELISA – SPD | Biovendor, LLC | Cat# RD194059101 |
| Luminex 10-plex (IL6, IL8, IL10, RAGE, Ang-2, ST2, TNFR1, fractalkine, procalcitonin, pentraxin3) | R&D Systems | Custom made: https://www.rndsystems.com/luminex/analytes |
| Karius McfDna-Seq | Karius Inc. | N/A |
| Deposited data | ||
| Karius mcfDNA results | Karius Inc. | Github repository: https://github.com/MicrobiomeALIR/Covid_mcfDNA.git |
| Software and algorithms | ||
| R version 3.5.1 | The R Foundation for Statistical Computing | N/A |
| Prism 9 | GraphPad | N/A |
| Other | ||
| Primary Code in R language and Datasets | Github | https://github.com/MicrobiomeALIR/Covid_mcfDNA.git |
| Primary Code in R language and Datasets | Zenodo | https://doi.org/10.5281/zenodo.8319850 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Kitsios (kitsiosg@upmc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw sequencing data collected for the study cannot be deposited in a public repository due to subject privacy, consent considerations and the proprietary nature of the Karius Test, factors that pose constraints related to raw data accessibility. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. We provided all sequencing results in detailed tables in the supplemental data items. In addition, Primary code and de-identified data for replication of analyses have been deposited at Github repository (https://github.com/MicrobiomeALIR/Covid_mcfDNA) and are publicly available as of the date of publication. DOIs are listed in the key resources table.
Experimental model and study participant details
Human subjects
From April 2020 through September 2020, following permission by the treating clinicians, we approached consecutive inpatient adults with acute hypoxemic respiratory failure and confirmed COVID-19 pneumonia, for enrollment to an observational, prospective cohort study. The University of Pittsburgh Institutional Review Board (IRB) approved the protocols and we obtained written or electronic informed consent by all participants or their surrogates in accordance with the Declaration of Helsinki. We enrolled critically ill patients from the ICU (n = 36) and moderately ill inpatients who were admitted in dedicated hospital wards (n = 6). We diagnosed COVID-19 pneumonia based on institutional clinical criteria (clinical symptoms, hypoxemia and abnormal chest radiographic findings) with confirmatory molecular testing (positive SARS-CoV-2 nasopharyngeal quantitative polymerase chain reaction [qPCR]). Patients were excluded if they were incarcerated or if they were receiving comfort-measures only care. We recorded history of pre-existing immunocompromising conditions upon enrollment (Table 1). We recorded glucocorticoid use for treatment of COVID-19, as well as the levels of respiratory support (invasive mechanical ventilation or ECMO) provided to the patients at the time of their enrollment. Given the exploratory design of our study and assembly of a feasibility dataset, we did not conduct an a priori sample size estimation.
Ethics approval and consent to participate
The University of Pittsburgh Institutional Review Board (IRB) approved the protocols (STUDY19050099 for critically ill patients, and STUDY20040036 for moderately ill inpatients). We obtained written or electronic informed consent by all participants or their surrogates in accordance with the Declaration of Helsinki.
Consent for publication
We obtained necessary patient/participant consent and the appropriate institutional forms have been archived. Any patient/participant/sample identifiers included were not known to anyone outside the research group so cannot be used to identify individuals.
Study group definitions
We reviewed all available clinical, microbiologic and antimicrobial treatment data around the timing of baseline samples (+/− 3 days) for patients with COVID-19 to ascertain the presence/absence or clinical suspicion for SI. We classified subjects in three groups, assessed by two reviewers.
-
a.
Microbiologically-Confirmed Secondary Infection (Micro-SI), when typical pathogenic microbes were isolated on clinical biospecimen cultures (blood, LRT, urine or tissue) and the subject was receiving antimicrobials for the documented infection.
-
b.
Clinically Diagnosed Secondary Infections (Clinical-SI), when empiric antimicrobials were administered without microbiologic SI confirmation.
-
c.
“No Clinical Suspicion for SI” (No-Suspected-SI), when microbiologic workup for SI was negative or not performed, and no empiric antimicrobials were prescribed.
We repeated classifications into the Micro-SI, Clinical-SI and No-Suspected-SI categories at the timing of follow-up samples (days 5 and 10 post-enrollment) from updated clinical and microbiologic data, when available, due to the dynamic nature of incident SIs and the corresponding changes in management by the treating clinicians. We quantified radiographic edema of baseline chest X-rays with the Radiographic Assessment of Lung Edema (RALE) score with the use of the PulmAnnotator software.27,36
Method details
Research sample collection
We collected blood samples in EDTA tubes on enrollment (baseline - day 1) for centrifugation, separation and storage of plasma and additional blood constituents until conduct of experiments. We also collected repeat blood samples on days 5 and 10 post-enrollment from critically ill patients who remained hospitalized in the intensive care unit (ICU), including extended follow-up if patients remained critically ill. From patients on invasive mechanical ventilation (IMV), we collected non-invasive endotracheal aspirates by gentle suctioning of an in-line suction catheter through the endotracheal tube with a standardized protocol.26
Laboratory analyses
Cell-free DNA assays
We conducted plasma microbial cell-free DNA metagenomic sequencing (mcfDNA-Seq) with the Karius Test [Karius Inc., Redwood City, CA] and classified the derived metagenomic sequences as human (hcfDNA) vs. microbial (mcfDNA). Based on minimum sequencing coverage metric required for quality control, we classified sequencing runs as “Pass”, “Qualitatively Pass” or “Failed”. Because the amount of hcfDNA in an individual’s blood can vary up to 1000-fold, the Karius Test utilizes a sample-specific minimum sequencing coverage metric for quality control that accounts for variation in the amount of hcfDNA by demanding more sequencing reads for those samples with more hcfDNA.21 Each specimen must receive a minimum number of internal control reads from the WINC (Whole-assay Internal Normalization Control) spike-in to pass the quality control threshold, thereby ensuring a more similar level of detection for each specimen regardless of hcfDNA background. On average, samples receive 20–25 million sequencing reads each, with about 10-fold variation depending on the concentration of background human hcfDNA in the sample. All samples must have at least 25,000 unique reads from the WINC spike-in to pass quality control.
The Karius Test involves several mitigation strategies for environmental contamination, which can lead to spurious results if not addressed. The contamination mitigation strategies include: (1) vendor selection for consumables and reagents that are assessed to be “DNA-free”, (2) novel, patented contamination-reduction processes that are used to decontaminate every reagent prior to use, (3) inclusion of four distinct environmental contamination control samples in every sequencing batch, with randomized positioning on the plate run, with reporting of only microbes whose presence cannot be explained by normal variation in the background levels of contamination (at a p value less than 10−50), (4) rigorous reagent qualification processes that screen every new shipment and lot of every critical reagent for changes in the environmental contamination profile, (5) internally designed assay controls to fail if any of the most common environmental contaminants are introduced unexpectedly, (6) computational surveillance of all results reported every seven days to detect statistical anomalies in test results that might be explained by contamination of a reagent. With these mitigation strategies, our approach collectively reduces the risk of false positives from environmental contamination by more than 10,000-fold.21
In addition to the sample-specific sequencing depth metric, the following quality control metrics are necessary for each sample and microbe to be reported: (1) The Assay Control samples for the batch must accurately report all spiked-in pathogens, at both high and low concentrations, without reporting any other pathogens; (2) every microbe on the test report must pass a Species Purity Ratio metric designed to assess whether there is any risk that the presence of a microbe in that specimen could be a result of cross-contamination from any other sample run in the last week; (3) no signs of sample mix-up based on unique molecular identifiers added to each specimen upon sample receipt; (4) there must be no statistical anomalies suggesting risk of false positives based on computational monitoring of all test reports within the past 7 days.
All microbes were reported at species level and included a quantitative measure of abundance expressed as DNA molecules per microliter (MPMs), with the exception of the “Qualitative Pass” calls where the reported organism could not be quantified. Healthy subjects have undetectable (77.2%) or very low mcfDNA levels, typically belonging to commensal organisms such as Helicobacter pylori, with DNA molecules per microliter (MPM) load averages ranging 2.64–4.05 MPM.20,21 We classified microbes identified by mcfDNA-Seq into recognized respiratory pathogens vs. microbes of unclear clinical importance based on systematic assessment of the literature implicating different organisms in lower respiratory tract infections (see Tables S5, S6 for detailed classifications). For contextualization, we compared mcfDNA levels among subjects with COVID-19 with our previously published dataset of mechanically ventilated patients with and without pneumonia.24 McfDNA-Seq analysis was performed as part of this research protocol, results were not available in real-time and not disclosed to the treating clinicians.
Bacterial and viral load assays
From available endotracheal aspirate samples collected concurrently with blood samples, we separately extracted genomic DNA and RNA and performed bacterial 16S rRNA gene and SARS-CoV-2 RNA qPCR for assessment of bacterial and viral load in the LRT, respectively.26,34 We measured plasma SARS-CoV-2 viral levels (vRNA) following RNA extraction and qPCR amplification with a sensitive in-house method.26
Host-response assays
From stored plasma samples, we measured baseline plasma levels of nine prognostic biomarkers of tissue injury and inflammation. We measured surfactant Protein D [SPD] with ELISA and the remainder eight biomarkers (interleukin [IL]-6, IL-8, pentraxin-3, procalcitonin, receptor for advanced glycation end products [RAGE], angiopoietin-2 (Ang-2), suppression of tumorigenicity [ST]-2, and tumor necrosis factor receptor [TNFR]-1) with a custom Luminex multi-analyte panel (R&D Systems, Minneapolis, MN, United States).
Quantification and statistical analysis
Statistical analyses
We compared clinical variables, biomarker and cfDNA levels between different categories (i.e., sequencing run success, SI clinical groups) and the historical non-COVID-19 cohort using non-parametric tests, Wilcoxon signed-rank and Fischer’s exact tests. We examined for correlations between biomarkers and cfDNA levels with the Pearson’s method, adjusted for multiple comparisons with the Benjamini-Hochberg method. We analyzed the impact of baseline mcfDNA levels on 90-day survival by Kaplan-Meier curves stratified by total mcfDNA tertiles and by constructing Cox proportional hazards models for log-10 transformed mcfDNA levels, adjusted for age, SI diagnosis, and inclusive of an interaction term for SI-mcfDNA. We analyzed survival at 90 days in all patients with baseline data (Figure 1). All SI categorizations represent the enrollment classification unless otherwise specified. No comparisons were made across follow-up periods (Day 1 vs. Day 5) unless otherwise specified. For “Pass” samples with zero microbial calls, we assigned a microbial MPM of “1” to allow for log10-transformations, and utilized log10-transformed values of plasma biomarkers and mcfDNA levels for analyses due to non-normal data.
Power analysis
We did not conduct an a priori sample size estimation due to the exploratory design of our study analyzing a feasibility dataset.
Acknowledgments
The authors wish to thank the patients and patient families that have enrolled in the University of Pittsburgh Acute Lung Injury Registry. We also thank the physicians, nurses, respiratory therapists and other staff at the University of Pittsburgh Medical Center Presbyterian, Shadyside and East Hospitals intensive care units for assistance with coordination of patient enrollment and collection of patient samples. We would also like to thank the laboratory personnel at the Center for Medicine and the Microbiome at the University of Pittsburgh for assistance with processing clinical samples.
Funding information: Dr. Kitsios: University of Pittsburgh Clinical and Translational Science Institute, COVID-19 Pilot Award; NIH (K23 HL139987; R03 HL162655). Dr. Bain: Career Development award number IK2 BX004886 from the U.S. Department of Veterans Affairs Biomedical Laboratory R&D (BLRD) Service. Dr. McVerry: NIH (P01 HL114453). The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report or decision to submit the manuscript for publication.
Author contributions
G.L.: conceptualization, methodology, validation, investigation, writing - original draft, writing - review and editing, visualization, project administration; R.D., A.A.A., T.B., S.B.: methodology, resources, investigation, writing - review and editing. M.H., N.A.Y., M.L., W.B., F.S., C.S., S.Q., X.W., Y.Z., J.L.J., A.N., G.H., J.W.M., B.M., B.J.M., A.M.: investigation, writing - review and editing; K.J.M., E.K.H.: software, resources, data curation, writing - review and editing; G.D.K.: conceptualization, methodology, validation, formal analysis, investigation, resources, writing - original draft, writing - review and editing, visualization, supervision, project administration, funding acquisition.
Declaration of interests
Drs. Duttagupta and Ahmed were employed by Karius, Inc at the time of the study but are no longer employed by Karius, Inc. Drs. Kitsios and Haidar have received research funding from Karius, Inc. Drs. Kitsios and Morris have received research funding from Pfizer, Inc. Dr. Haidar serves on the Karius, Inc scientific advisory board. Dr. Mellors is a consultant to AlloVir, Infectious Disease Connect, Inc., and Gilead Sciences, Inc., has received research funding from Gilead Sciences, Inc. to the University of Pittsburgh, receives compensation from Abound Bio, Inc. (unrelated to the current work) and holds shares options in Galapogos, Infectious Disease Connect, Inc., and MingMed Biotechnology Co. Ltd. (unrelated to the current work). Dr. McVerry has received research funding from Bayer Pharmaceuticals, Inc. and consulting fees from Boehringer Ingelheim, both unrelated to this work.
Published: October 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108093.
Supplemental information
Abbreviations: Not detected (ND); no growth (NG); normal respiratory flora (NRF); gram stain (GS); culture (Cx); gram negative rods (GNRs); gram positive cocci in chains (GPCCH); gram positive cocci in pairs (GPCP); T2 Candida fungal test (T2).
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouzé A., Martin-Loeches I., Povoa P., Makris D., Artigas A., Bouchereau M., Lambiotte F., Metzelard M., Cuchet P., Boulle Geronimi C., et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: a European multicenter cohort study. Intensive Care Med. 2021;47:188–198. doi: 10.1007/s00134-020-06323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maes M., Higginson E., Pereira-Dias J., Curran M.D., Parmar S., Khokhar F., Cuchet-Lourenço D., Lux J., Sharma-Hajela S., Ravenhill B., et al. Ventilator-associated pneumonia in critically ill patients with COVID-19. Crit. Care. 2021;25:25. doi: 10.1186/s13054-021-03460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickens C.O., Gao C.A., Cuttica M.J., Smith S.B., Pesce L.L., Grant R.A., Kang M., Morales-Nebreda L., Bavishi A.A., Arnold J.M., et al. Bacterial Superinfection Pneumonia in Patients Mechanically Ventilated for COVID-19 Pneumonia. Am. J. Respir. Crit. Care Med. 2021;204:921–932. doi: 10.1164/rccm.202106-1354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitsios G.D., Morris A. Seek and Ye Shall Find: COVID-19 and Bacterial Superinfection. Am. J. Respir. Crit. Care Med. 2021;204:875–877. doi: 10.1164/rccm.202107-1790ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivram H., Hackney J.A., Rosenberger C.M., Teterina A., Qamra A., Onabajo O., McBride J., Cai F., Bao M., Tsai L., et al. Tocilizumab treatment leads to early resolution of myeloid dysfunction and lymphopenia in patients hospitalized with COVID-19. bioRxiv. 2022 doi: 10.1101/2022.10.27.514096. Preprint at. [DOI] [Google Scholar]
- 7.Conway Morris A., Kohler K., De Corte T., Ercole A., De Grooth H.J., Elbers P.W.G., Povoa P., Morais R., Koulenti D., Jog S., et al. Co-infection and ICU-acquired infection in COIVD-19 ICU patients: a secondary analysis of the UNITE-COVID data set. Crit. Care. 2022;26:236. doi: 10.1186/s13054-022-04108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng J., Fu M., Mei H., Zheng H., Liang G., She X., Wang Q., Liu W. Efficacy and secondary infection risk of tocilizumab, sarilumab and anakinra in COVID-19 patients: A systematic review and meta-analysis. Rev. Med. Virol. 2022;32:e2295. doi: 10.1002/rmv.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes S., Troise O., Donaldson H., Mughal N., Moore L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020;26:1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C.A., Bailey J.I., Walter J.M., Coleman J.M., Malsin E.S., Argento A.C., Prickett M.H., Wunderink R.G., Smith S.B. Bronchoscopy on intubated patients with COVID-19 is associated with low infectious risk to operators. Ann. Am. Thorac. Soc. 2021;18:1243–1246. doi: 10.1513/AnnalsATS.202009-1225RL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giron L.B., Dweep H., Yin X., Wang H., Damra M., Goldman A.R., Gorman N., Palmer C.S., Tang H.Y., Shaikh M.W., et al. Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paludan Søren T.M., Mogensen T.H. Innate immunological pathways in COVID-19 pathogenesis. Sci. Immunol. 2022;7:eabm5505. doi: 10.1126/sciimmunol.abm5505. [DOI] [PubMed] [Google Scholar]
- 13.Abers M.S., Delmonte O.M., Ricotta E.E., Fintzi J., Fink D.L., de Jesus A.A.A., Zarember K.A., Alehashemi S., Oikonomou V., Desai J.V., et al. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6 doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heer R.S., Mandal A.K., Kho J., Szawarski P., Csabi P., Grenshaw D., Walker I.A., Missouris C.G. Elevated procalcitonin concentrations in severe Covid-19 may not reflect bacterial co-infection. Ann. Clin. Biochem. 2021;58:520–527. doi: 10.1177/00045632211022380. [DOI] [PubMed] [Google Scholar]
- 15.Schuurman A.R., Reijnders T.D.Y., van Engelen T.S.R., Léopold V., de Brabander J., van Linge C., Schinkel M., Pereverzeva L., Haak B.W., Brands X., et al. The host response in different aetiologies of community-acquired pneumonia. EBioMedicine. 2022;81 doi: 10.1016/j.ebiom.2022.104082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Togashi Y., Kono Y., Okuma T., Shioiri N., Mizushima R., Tanaka A., Ishiwari M., Toriyama K., Kikuchi R., Takoi H., Abe S. Surfactant protein D: A useful biomarker for distinguishing COVID-19 pneumonia from COVID-19 pneumonia-like diseases. Health Sci. Rep. 2022;5:e622. doi: 10.1002/hsr2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen A.J., Glick L.R., Lee S., Kunitomo Y., Tsang D.A., Pitafi S., Valda Toro P., Ristic N.R., Zhang E., Carey G.B., et al. Nonutility of procalcitonin for diagnosing bacterial pneumonia in patients with severe COVID-19. Eur. Clin. Respir. J. 2023;10 doi: 10.1080/20018525.2023.2174640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langford B.J., So M., Raybardhan S., Leung V., Soucy J.P.R., Westwood D., Daneman N., MacFadden D.R. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infect. 2021;27:520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell C.D., Fairfield C.J., Drake T.M., Turtle L., Seaton R.A., Wootton D.G., Sigfrid L., Harrison E.M., Docherty A.B., de Silva T.I., et al. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. Lancet. Microbe. 2021;2:e354–e365. doi: 10.1016/s2666-5247(21)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalantar K.L., Neyton L., Abdelghany M., Mick E., Jauregui A., Caldera S., Serpa P.H., Ghale R., Albright J., Sarma A., et al. Integrated host-microbe plasma metagenomics for sepsis diagnosis in a prospective cohort of critically ill adults. Nat. Microbiol. 2022;7:1805–1816. doi: 10.1038/s41564-022-01237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blauwkamp T.A., Thair S., Rosen M.J., Blair L., Lindner M.S., Vilfan I.D., Kawli T., Christians F.C., Venkatasubrahmanyam S., Wall G.D., et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019;4:663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 22.Kitsios G.D., Bain W., Al-Yousif N., Duttagupta R., Ahmed A.A., McVerry B.J., Morris A. Plasma microbial cell-free DNA load is associated with mortality in patients with COVID-19. Respir. Res. 2021;22:24. doi: 10.1186/s12931-021-01623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer A., Buetti N., Houhou-Fidouh N., Patrier J., Abdel-Nabey M., Jaquet P., Presente S., Girard T., Sayagh F., Ruckly S., et al. HSV-1 reactivation is associated with an increased risk of mortality and pneumonia in critically ill COVID-19 patients. Crit. Care. 2021;25:417. doi: 10.1186/s13054-021-03843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H., Haidar G., Al-Yousif N.S., Zia H., Kotok D., Ahmed A.A., Blair L., Dalai S., Bercovici S., Ho C., et al. Circulating microbial cell-free DNA is associated with inflammatory host-responses in severe pneumonia. Thorax. 2021;76:1231–1235. doi: 10.1136/thoraxjnl-2020-216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs J.L., Bain W., Naqvi A., Staines B., Castanha P.M.S., Yang H., Boltz V.F., Barratt-Boyes S., Marques E.T.A., Mitchell S.L., et al. SARS-CoV-2 Viremia is Associated with COVID-19 Severity and Predicts Clinical Outcomes. Clin. Infect. Dis. 2022;74:1525–1533. doi: 10.1093/cid/ciab686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs J.L., Naqvi A., Shah F.A., Boltz V.F., Kearney M.F., McVerry B.J., Ray P., Schaefer C., Fitzpatrick M., Methé B., et al. Plasma SARS-CoV-2 RNA Levels as a Biomarker of Lower Respiratory Tract SARS-CoV-2 Infection in Critically Ill Patients With COVID-19. J. Infect. Dis. 2022;226:2089–2094. doi: 10.1093/infdis/jiac157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Yousif N., Komanduri S., Qurashi H., Korzhuk A., Lawal H.O., Abourizk N., Schaefer C., Mitchell K.J., Dietz C.M., Hughes E.K., et al. Inter-rater reliability and prognostic value of baseline Radiographic Assessment of Lung Edema (RALE) scores in observational cohort studies of inpatients with COVID-19. BMJ Open. 2023;13 doi: 10.1136/bmjopen-2022-066626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andargie T.E., Tsuji N., Seifuddin F., Jang M.K., Yuen P.S., Kong H., Tunc I., Singh K., Charya A., Wilkins K., et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight. 2021;6 doi: 10.1172/jci.insight.147610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atallah N.J., Warren H.M., Roberts M.B., Elshaboury R.H., Bidell M.R., Gandhi R.G., Adamsick M., Ibrahim M.K., Sood R., Bou Zein Eddine S., et al. Baseline procalcitonin as a predictor of bacterial infection and clinical outcomes in COVID-19: A case-control study. PLoS One. 2022;17 doi: 10.1371/journal.pone.0262342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz M., Maiberger T., Armbrust L., Kiwit A., Von der Wense A., Reinshagen K., Elrod J., Boettcher M. cfDNA and DNases: New Biomarkers of Sepsis in Preterm Neonates-A Pilot Study. Cells. 2022;11:192. doi: 10.3390/cells11020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing Q., Leung C.H.C., Wu A.R. Cell-Free DNA as Biomarker for Sepsis by Integration of Microbial and Host Information. Clin. Chem. 2022;68:1184–1195. doi: 10.1093/clinchem/hvac097. [DOI] [PubMed] [Google Scholar]
- 32.Savva A., Roger T. Targeting toll-like receptors: promising therapeutic strategies for the management of sepsis-associated pathology and infectious diseases. Front. Immunol. 2013;4:387. doi: 10.3389/fimmu.2013.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvioni L., Testa F., Sulejmani A., Pepe F., Giorgio Lovaglio P., Berta P., Dominici R., Leoni V., Prosperi D., Vittadini G., et al. Surfactant protein D (SP-D) as a biomarker of SARS-CoV-2 infection. Clin. Chim. Acta. 2022;537:140–145. doi: 10.1016/j.cca.2022.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitsios G.D., Nguyen V.D., Sayed K., Al-Yousif N., Schaefer C., Shah F.A., Bain W., Yang H., Fitch A., Li K., et al. The upper and lower respiratory tract microbiome in severe aspiration pneumonia. iScience. 2023;26 doi: 10.1016/j.isci.2023.106832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K.H., Won D., Kim J., Lee J.A., Kim C.H., Kim J.H., Jeong S.J., Ku N.S., Choi J.Y., Yeom J.-S., et al. Utility of Plasma Microbial Cell-Free DNA Whole-Genome Sequencing for Diagnosis of Invasive Aspergillosis in Patients With Hematologic Malignancy or COVID-19. J. Infect. Dis. 2023;228:444–452. doi: 10.1093/infdis/jiad213. [DOI] [PubMed] [Google Scholar]
- 36.Kotok D., Yang L., Evankovich J.W., Bain W., Dunlap D.G., Shah F., Zhang Y., Manatakis D.V., Benos P.V., Barbash I.J., et al. The evolution of radiographic edema in ARDS and its association with clinical outcomes: A prospective cohort study in adult patients. J. Crit. Care. 2020;56:222–228. doi: 10.1016/j.jcrc.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abbreviations: Not detected (ND); no growth (NG); normal respiratory flora (NRF); gram stain (GS); culture (Cx); gram negative rods (GNRs); gram positive cocci in chains (GPCCH); gram positive cocci in pairs (GPCP); T2 Candida fungal test (T2).
Data Availability Statement
-
•
Raw sequencing data collected for the study cannot be deposited in a public repository due to subject privacy, consent considerations and the proprietary nature of the Karius Test, factors that pose constraints related to raw data accessibility. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. We provided all sequencing results in detailed tables in the supplemental data items. In addition, Primary code and de-identified data for replication of analyses have been deposited at Github repository (https://github.com/MicrobiomeALIR/Covid_mcfDNA) and are publicly available as of the date of publication. DOIs are listed in the key resources table.