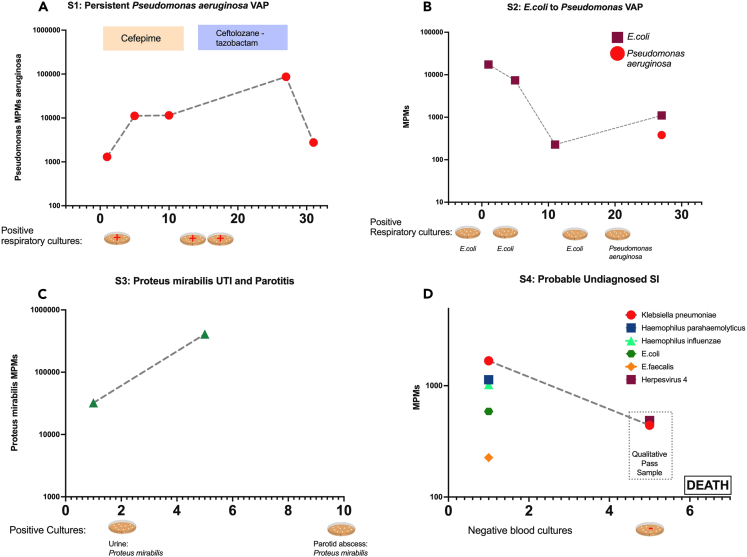
Figure 4.
Comparison of plasma mcfDNA sequencing with microbiologic/clinical diagnoses across the timeline of four subjects with serial sampling
Species-specific microbial MPMs (y axis) are shown across sequential sampling days from enrollment, (x axis). Petri dish graphics along the x axis denote the timing and results of clinically obtained microbiological testing. Subject 3 (A) was an immunocompetent patient with a persistent culture-confirmed Pseudomonas VAP that persisted through the antimicrobials, with timing and therapy noted above. Subject 3 had persistently elevated Pseudomonas mcfDNA levels throughout the extended infection course, which ultimately correlated with clinical improvement (A). Subject 4 (B) had sequential VAPs with changing pathogens detected on invasive respiratory cultures, E. coli followed by Pseudomonas, which was concordantly reported on noninvasive mcfDNA sampling. Subject 6 (C) was a patient with diabetes who was initially found to have a resistant Proteus urinary infection, and a subsequent persistent septic clinical picture, later found to have a polymicrobial, including Proteus, parotid gland abscess. Proteus mcfDNA levels remained elevated in subject 6 during initial antimicrobial therapy for urinary infection, suggesting the persistent source of infection. Subject 26 (D) was clinically determined No-Suspicion-for-SI, but had a deteriorating clinical course, without cultures obtained for 5 days or empiric antimicrobials, and ultimately died of shock and multisystem organ failure. Noninvasive testing revealed persistent levels of Klebsiella pneumonia mcfDNA levels, which suggests an undiagnosed SI may have contributed to the clinical course. See also Figure S7.