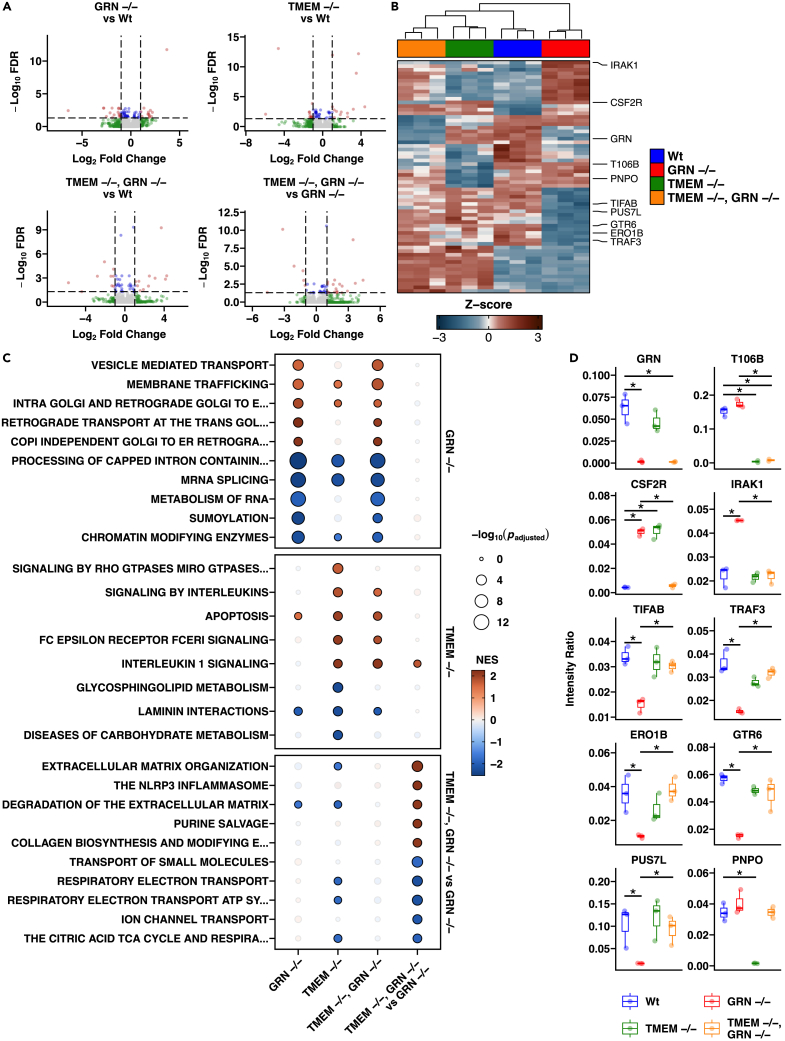
Figure 2.
TMEM106B and GRN KO iMG proteomic signatures
(A) Volcano plots of differential gene expression profiles from pairwise contrasts of GRN KO vs. WT, TMEM KO vs. WT, double KO vs. WT, and double KO vs. GRN KO. Red indicates proteins passing the FDR <0.05 & |Log2 FC| > 1 threshold, blue indicates proteins passing the FDR <0.05 threshold, green represents proteins passing the |Log2 FC| > 1 threshold, and gray represents the remaining proteins.
(B) Heatmap of significant (FDR <0.05 & |Log2 FC| > 1) differentially expressed proteins presented in the volcano plots.
(C) Enrichment testing results for the top distinct significant (padjusted < 0.05) pathways for GRN KO vs. WT, TMEM KO vs. WT, and double KO vs. GRN KO. Values for double KO vs. WT are also shown.
(D) Expression of select significant (FDR <0.05 & |Log2 FC| > 1) differentially expressed proteins from pairwise contrasts of GRN KO vs. WT, TMEM KO vs. WT, double KO vs. WT, and double KO vs. GRN KO within each genotype. Error bars represent the S.E.M. for 3 biological replicates.