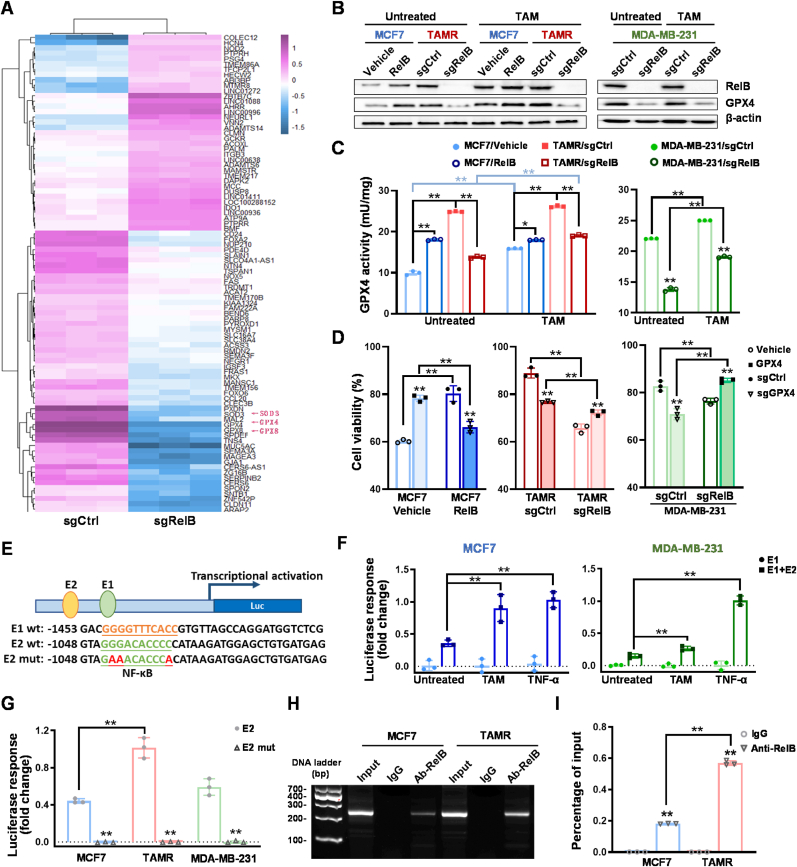
Fig. 7.
RelB-upregulated GPX4 expression. (A) The transcriptomes in RelB KO vs. control MDA-MB-231 cells were analyzed by RNA-seq. The antioxidants including GPX4 are indicated by red arrows. (B) After TAM treatment, the protein levels of GPX4 in the RelB-manipulated cells were quantified by immunoblotting. (C) The relative GPX4 activity in TAM-treated cells was measured (n = 3). (D) After TAM treatment, TAM-treated cytotoxicity was estimated by MTT assay (n = 3). (E) A 5′-flanking region of the human GPX4 gene containing two putative NF-κB elements (E1 and E2) with a core promoter was cloned to drive the luciferase reporter expression. The E1 and E2 sites were further mutated as indicated by red letters. (F) TNF-α- and TAM-induced enhancer responses in TAMS and TAMR cells were estimated by β-gal-normalized luciferase reporter activities (n = 3). (G) The E2 site for enhancer response was determined using its mutant construct (n = 3). (H, I) Furthermore, the E2 site was precipitated using a RelB antibody and quantified by standard PCR (H) and qPCR (I) (n = 3). Chromatins without immunoprecipitation served as an input control, and IgG-precipitated chromatins served as a negative control. Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01), two-way ANOVA. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)