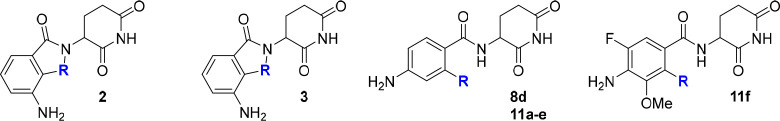
Table 2. Chemical Structures, Binding Data, Physicochemical Properties, and Cellular Activities of Ortho-Substituted Benzamides.
| neosubstrate
deg (%)h,i |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | R | IC50 (μM)a | Ki (μM)a | elog D7.4b | PPB (%)c | CHIIAMd | log(S)e | Redox activityf | UV/vis stabilityg | IKZF3 | SALL4 |
| 2 | C=O | 13 ± 2.7 | 3.3 ± 1.4 | 0.5 | 51 | –4.0 | –4.0 | n.a.j | <pH 9 | 86 | 95 |
| 3 | CH2 | 19 ± 1.5 | 6.4 ± 0.8 | –0.4 | 12 | –2.6 | –2.6 | not active | stable | 57 | 70 |
| 8d | F | 63 ± 16 | 29 ± 8.2 | –0.7 | 5 | 5.2 | –3.1 | not active | stable | 39 | 14 |
| 11a | Cl | 60 ± 13 | 28 ± 6.6 | –0.9 | 10 | 2.6 | –2.6 | not active | stable | 13 | <5 |
| 11b | CF3 | 87 ± 25 | 42 ± 13 | –0.3 | 6 | 6.7 | –3.5 | not active | stable | 69 | 45 |
| 11c | CH3 | 132 ± 55 | 65 ± 29 | –1.2 | 2 | –1.1 | –2.5 | not active | stable | 57 | <5 |
| 11d | OMe | 28 ± 2.6 | 11 ± 1.4 | 0.0 | 17 | 6.5 | –2.9 | not active | stable | 14 | 14 |
| 11e | OH | 20 ± 2.0 | 6.8 ± 1.0 | –0.3 | 18 | 8.8 | –3.0 | active | <pH 9 | 41 | <5 |
| 11f | F | 90 ± 17 | 44 ± 9.0 | –0.9 | 14 | 2.6 | –3.0 | not active | stable | 63 | 42 |
Affinity values determined in a competitive MST assay as described in the method sections (see Supporting Information).
Distribution coefficients at pH 7.4 were estimated by an HPLC-based method.
Plasma protein binding; experimentally determined percentage of compound bound to human serum albumin.
Chromatographic hydrophobicity index values referring to IAM chromatography (CHIIAM values), an estimate for drug–membrane interactions and permeability.
Logarithm of the solubility measured in mol/L at pH 6.8 by an HPLC-based method.
Redox activity assays for the detection of compounds that react with reducing agents in redox cycles by forming ROS (H2DCFDA assay) or free radicals (resazurin assay); see ref (49).
UV–vis-based assay for the evaluation of aqueous stability in phosphate buffer at pH 7.0, 8.0, and 9.0 after 4 h of incubation at 37 °C.
Percentage of degraded IKZF3 protein after 16 h treatment of MM.1S cells with 0.1 μM of each compound.
Percentage of degraded SALL4 protein after 16 h treatment of HuH6 cells with 0.1 μM of each compound. Western blots were analyzed by densitometric methods, and values were normalized to the respective loading controls and to DMSO-treated conditions. IKZF3/SALL4 degradation data represent the average of at least two independent biological experiments.
Not available due to spectral interference.