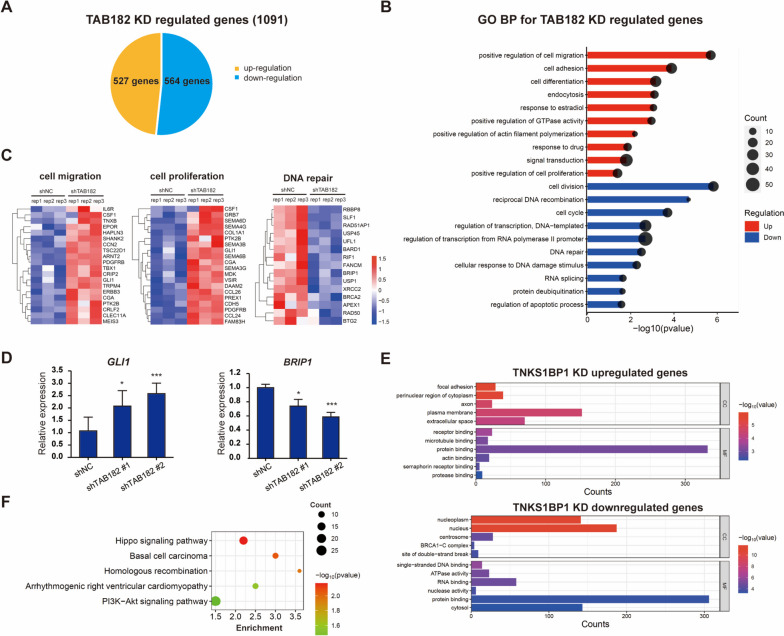
Fig. 3.
TAB182 regulated global gene expression in TNBC cells. A The pie chart presents the numbers of TAB182 KD-regulated genes (|Fold change|> 1.5, P < 0.05). B DAVID functional annotation shows the top 10 significant gene ontology biological processes (GO BP) for genes that were highly upregulated (upper) and downregulated (down) after TAB182 knockdown in MDA-MB-231 cells. P < 0.05. C Heatmaps showing the expression of TAB182 deletion-regulated genes enriched in three BP terms, including cell migration, cell proliferation, and DNA repair. The data are presented as transformed FPKM values of shTAB182 vs. shNC experiments. D A RT‒qPCR analysis confirmed the changes in gene expression identified by RNA-seq assay after TAB182 deletion. The data are shown as the means with SD relative to the control (shNC) from three independent experiments. * P < 0.05, *** P < 0.001 by Student’s t test. E GO analysis of cell component (CC) and molecular function (MF) of the genes upregulated and downregulated after TAB182 deletion, respectively. F KEGG pathway analysis [33] revealed the signaling pathways significant enriched by TAB182 KD-regulated genes in MDA-MB-231 cells