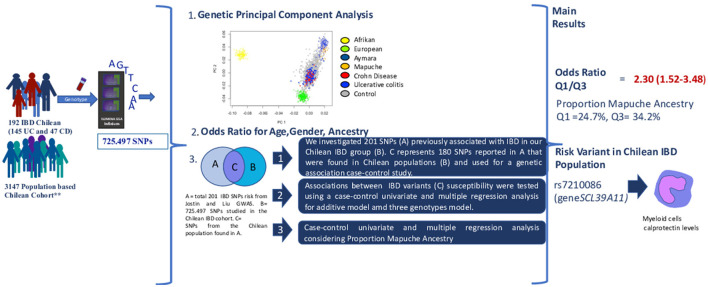
Figure 4.
Summary workflow of the study. 192 IBD Chilean patients were genotyped for 725497 SNP using Illumina GSA Infinium. Genotype data were combined with similar information from 3,147 Chilean controls **(30). (1) Proportions of Aymara, African, European, and Mapuche ancestry were estimated. (2) We calculated the odds ratios (OR) and 95% confidence intervals (CI) for gender, as well as age, and ancestry proportions. (3) We also explored associations with previously reported IBD-risk variants (8, 9) independently and in conjunction with genetic ancestry. Main results. The first and third quartiles of the proportion of Mapuche ancestry in IBD patients were 24.7 and 34.2%, respectively, and the corresponding OR was 2.30 (95% CI 1.52–3.48) for the lowest vs. the highest group. The risk variant rs7210086 related to myeloid cells was associated with IBD in the Chilean cohort (rs7210086-C, risk allele). IBD, Inflammatory bowel disease; SNPs, Single nucleotide polymorphism; GWAS, Genome wide analysis sequence; Q1, Quartile 1; Q2, Quartile 2.