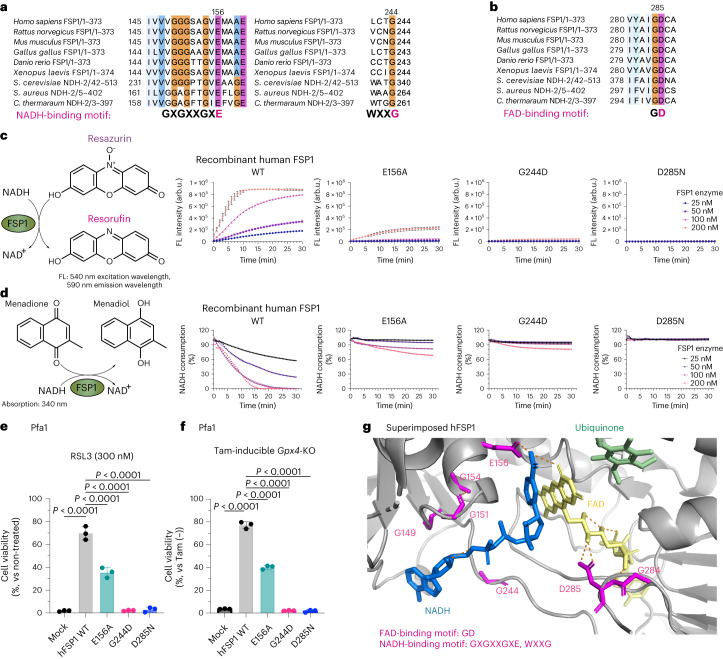
Fig. 1. G244 is essential for the predicted NAD(P)H-binding site of FSP1.
a, Conserved NADH-binding site (GXGXXGXE and WXXG). Protein alignment of FSP1 from different species: Homo sapiens (human), Mus musculus (mouse), Rattus norvegicus (rat), Gallus gallus (chicken), Xenopus laevis (frog), Danio rerio (zebrafish), Saccharomyces cerevisiae Ndi1 (PDB: 4G73), Caldalkalibacillus thermarum NDH-2 (PDB: 5NA1), and Staphylococcus aureus NDH-2 (PDB: 4NWZ). The numbers next to the organisms indicate the range of the amino acids of each gene used for generating the protein alignment. b, Conserved FAD-binding site (GD). Protein alignment of FSP1 from different species. c, Schematic representation of the FSP1 enzyme activity assay with resazurin used as the substrate (left). Reduction of resazurin in the presence of wild-type (WT) FSP1, FSP1-E156A, FSP1-G244D, or FSP1-D285N at the indicated concentrations. Data are shown as the mean ± s.d. of 3 wells of a 96-well plate, from 1 of 3 independent experiments. FL, fluorescence. d, Schematic representation of FSP1 enzyme activity assay, with menadione used as the substrate (left). Oxidation of NADH in the presence of WT FSP1, FSP1-E156A, FSP1-G244D, or FSP1-D285N at the indicated concentrations. Data are from a single well of a 96-well plate from 1 of 3 independent experiments. e, Viability of Pfa1 cells stably overexpressing WT HA-tagged human FSP1 (hFSP1-HA) or FSP1 mutants, treated with 300 nM RSL3 for 24 h. Data are shown as the mean ± s.d. of 3 wells of a 96-well plate from 1 of 3 independent experiments. f, Viability was measured in Pfa1 cells stably overexpressing WT hFSP1 or one of the mutants with or without treatment with Tam (1 µM) for 72 h. Data were normalized to each group that was not treated with Tam. Data are shown as the mean ± s.d. of 3 wells of a 96-well plate from 1 of 3 independent experiments. P values were calculated using a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple-comparison test (e,f). g, Predicted FSP1 structure, with consensus NADH- and FAD-binding motifs, from the AlphaFold2 database (https://alphafold.ebi.ac.uk). The co-factors, FAD (yellow), NADH (blue) and CoQ5 (green), were embedded from the structure of the yeast ortholog, NDH-2 (Ndi1) (PDB: 4G73). The expected hydrogen bond was generated by Pymol.