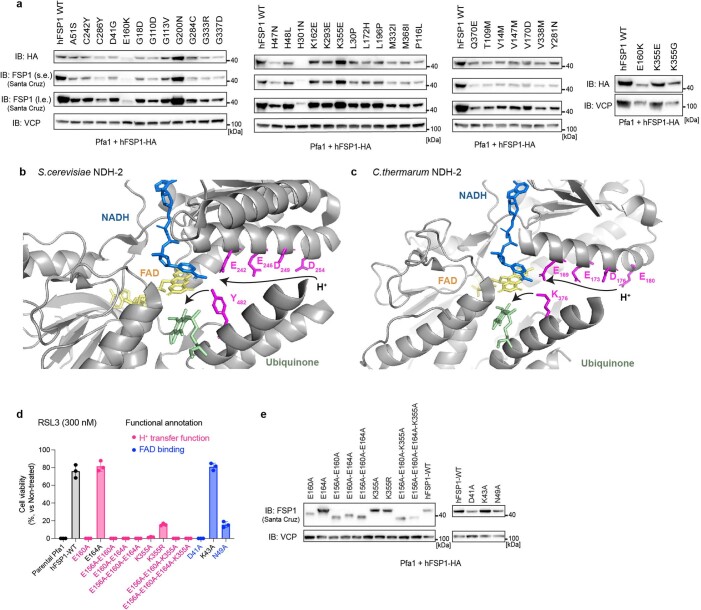
Extended Data Fig. 3. Dysfunctional FSP1 mutations and proposed proton transfer function in NDH-2s.
a. Immunoblot analysis of FSP1 (using AMID (Santa Cruz) and HA antibodies) and VCP expression in Pfa1 cells stably overexpressing wildtype hFSP1 or its mutant variants. For immunoblot of FSP1, s.e. and l.e. represent short and long exposure, respectively. b. Crystal structure of Saccharomyces cerevisiae (S. cerevisiae) NDH-2 (PDB: 4G73) with its cofactors, FAD (yellow), NADH (blue), and CoQ5 (green). The proton transfer via sequential carboxylic residues in the α-helix (that is, E242/E246/D249/D294) and subsequent final protonation from Y482 to ubiquinone are indicated with black arrows. c. Crystal structure of Caldalkalibacillus thermarum (C. thermarum) NDH-2 (PDB: 4NWZ) with its cofactors, FAD (yellow), NADH (blue), and CoQ5 (green). The proton transfer via sequential carboxylic residues in the α-helix (that is, E169/E173/D176/E180) and subsequent final protonation from K376 to ubiquinone are indicated with black arrows. d. Cell viability was measured after treating Pfa1 cells stably overexpressing wildtype hFSP1 or its mutant variants with or 300 nM RSL3 for 24 h. Data was normalized by each group of non-treatment with RSL3. Data represents the mean ± SD of 3 wells of a 96-well plate from one out of 3 independent experiments. e. Immunoblot analysis of FSP1 (Santa Cruz) and VCP expression in Pfa1 cells stably overexpressing wildtype hFSP1 or its mutant variants in cells. Data is shown from a single experiment (a,e).