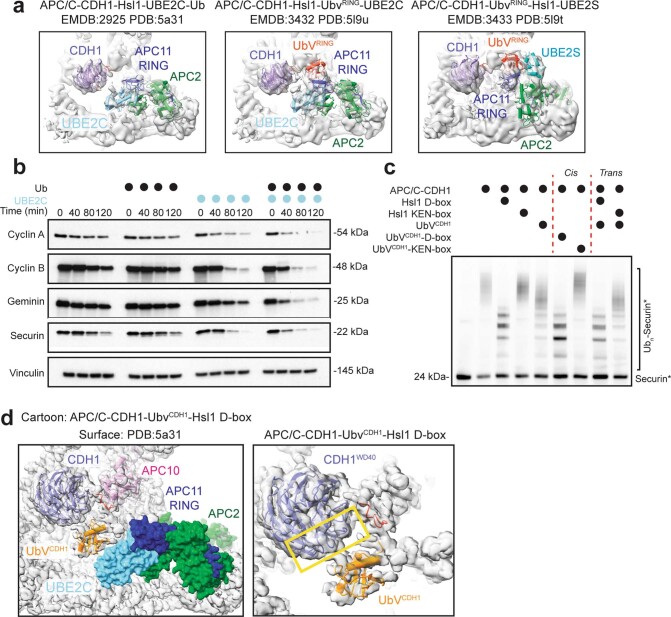
Extended Data Fig. 6. Previous artificially-trapped APC/C structures mimicking substrate ubiquitination.
a – Fitted cryoEM models from previous studies of the APC/C showing monoubiquitination (left), multi-ubiquitination (center), and polyubiquitination (right)23,38. An Hsl1 truncation harboring its KEN- and D-box was used as a substrate where a lysine was replaced with cysteine and crosslinked to the active site of UBE2C to mimic substrate priming (left). A similar Hsl1 truncation was genetically fused to the UbVRING, purified, and crosslinked to UBE2C and Ub to mimic multi-monoubiquitination (center). For chain elongation, a UbVRING-Hsl1 D-box chimera harboring a K11C substitution was crosslinked to the active site of UBE2S (right). b – Similar to Fig. 4e, immunoblotting of APC/C-dependent substrate degradation in mitotic HeLa cell extracts reveals that this process is not inhibited by the addition of excess Ub, in contrast to the UbVCDH1. The experiments were performed independently three times. c – Similar to Fig. 4f, Securin ubiquitination is inhibited more strongly by the UbVCDH1-Hsl1 D-box fusion, as monitored by fluorescent scanning of APC/C-CDH1-UBE2C-dependent ubiquitination reactions. The experiments were performed independently three times. d –Cryo-EM map of APC/C-CDH1-UbVCDH1-Hsl1 D-box rigid body fitted with a surface representation model of UBE2C bound to APC2 (green)/APC11(blue). Left, UbVCDH1 (orange) is in position to receive Ub from UBE2C (cyan). Right, cryo-EM map fitted with atomic models of CDH1 (purple) and UbVCDH1 (orange)-Hsl1 D-box (red). Yellow rectangle indicates the KEN-box binding site of CDH1, similar to where UbVCDH1 binds. Uncropped gels representative of n = 3 independent experiments for C available in source data.