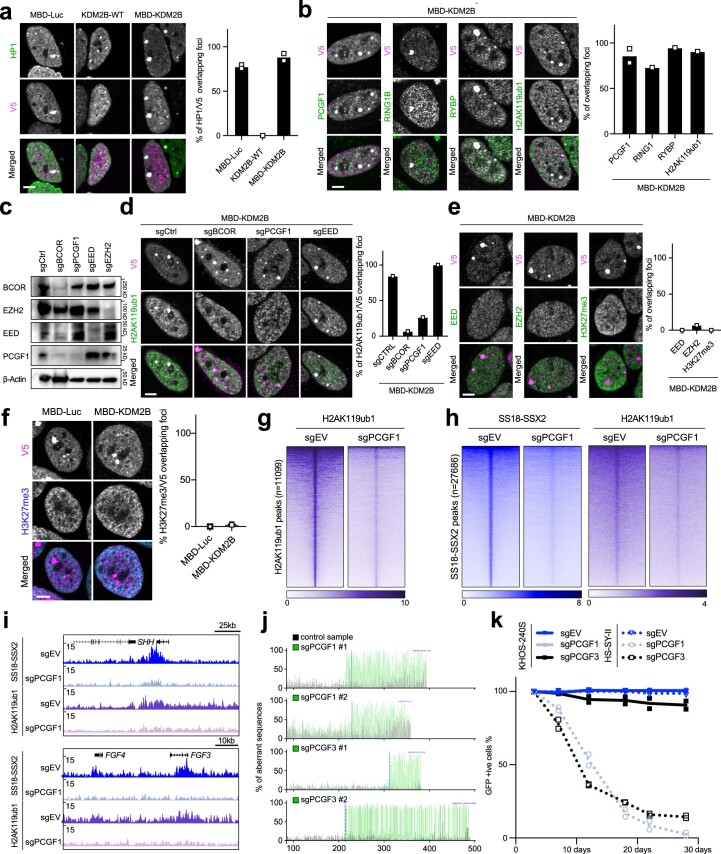
Extended Data Fig. 1. PRC1.1 is suficient to initiate SS18-SSX recruitment.
a) Left, Immunofluorescence for the MBD-Luc, MBD-KDM2B or for KDM2B-WT fused to a V5 tag (V5, magenta) and HP1 (green). Right, percentage of V5 foci overlapping HP1 foci. Data represents the mean of 2 biological replicates. Scale bars 5um throughout the figure. b) Left, Immunofluorescence of MBD-KDM2B (V5, magenta) with PCGF1, RING1B, RYBP and H2AK119ub1 (green). Right, percentage of foci overlapping a V5 foci in n = 2 (PCGF1, data represents the mean) or 1 biological replicate. c) Western Blot of HS-SY-II-Cas9 whole cell extracts expressing sgRNAs revealed using BCOR, EZH2, EED, PCGF1 or Beta-actin antibodies. d) Left, Immunofluorescence for MBD-KDM2B (V5, magenta) in the presence of different sgRNAs (resulting in eGFP background fluorescence) with H2AK119ub1 (green). Right, percentage of H2AK119ub1 foci overlapping V5 foci in one biological replicate. e) Left, Immunofluorescence of MBD-KDM2B (V5, magenta) with EED, EZH2 or H3K27me3 (green). Right, percentage of foci overlapping a V5 foci in one biological replicate. f) Left, Immunofluorescence for V5 (magenta) and H3K27me3 (cyan). Right, percentage of H3K27me3 foci overlapping with V5 foci in 2 biological replicates. Data represents the mean. g) Heatmaps of H2AK119ub1 scaled CUT&RUN signals (purple) in HS-SY-II-Cas9 cells expressing empty sgRNA as control (sgEV) or targeting PCGF1 (sgPCGF1) over H2AK119ub1 peaks in HS-SY-II (n = 11099). Rows correspond to ±10-kb regions across the midpoint of each enriched region, ranked by increasing signal. h) Heatmaps of SS18-SSX2 (blue) or H2AK119ub1 (purple) scaled CUT&RUN signals in SYO-I-Cas9 cells expressing empty sgRNA as control (sgEV) or targeting PCGF1 (sgPCGF1) over SS18-SSX2 peaks in SYO-I (n = 27686). Rows correspond to ±10-kb regions across the midpoint of each enriched region, ranked by increasing signal. i) Gene tracks for H2AK119ub1 and SS18-SSX CUT&RUN signals in SYO-I Cas9 at the SHH and FGF4-FGF3 loci. j) Tracking of Indels by Decomposition (TIDE) assay displaying the percentage of aberrant sequences after Cas9 editing for 2 guides targeting PCGF1 and PCGF3 versus the wild-type sequence (control sample). k) Cell competition assay performed in the osteosarcoma cell line KHOS-240S-Cas9 (fusion negative control) or in the synovial sarcoma line HS-SY-II-Cas9 transduced with an empty sgRNA as control or with guides targeting PCGF1 and PCGF3.