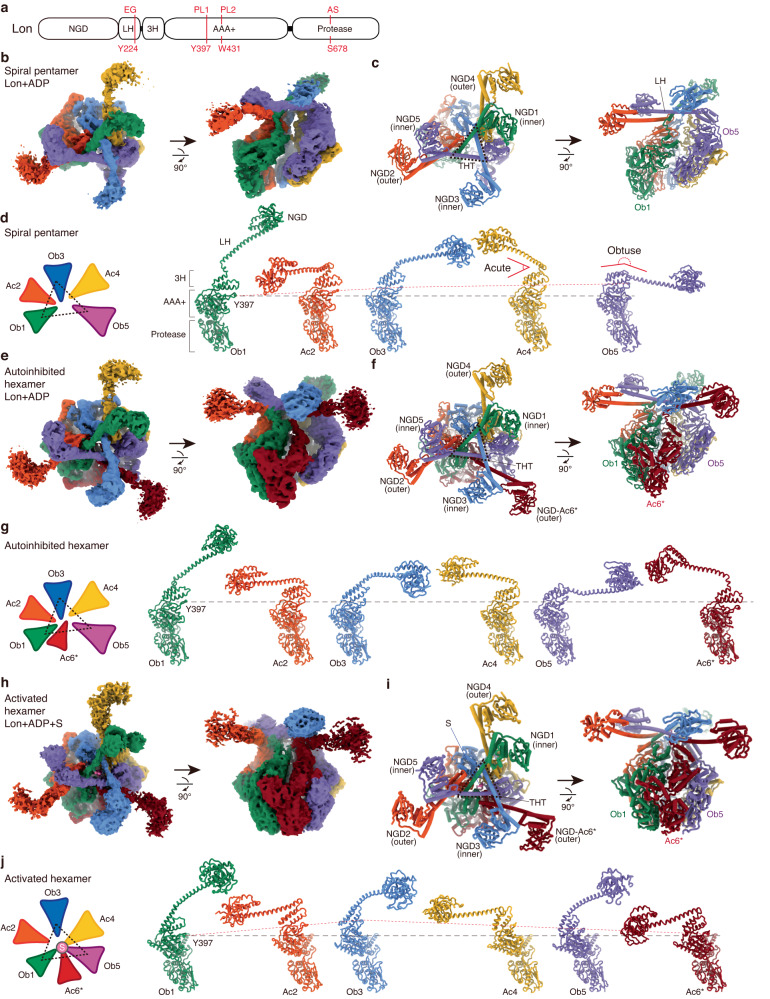
Fig. 1. Cryo-EM structures of Lon in three assembled states.
a Each Lon protomer consists of an N-terminal globular domain (NGD), a long helix (LH), a 3-helix (3H) bundle, a AAA+ ATPase domain, and a protease domain. The locations of the residues of the entry gate (EG), the pore loops (PL1 and PL2), and the proteolytic active site (AS) are indicated. b, c Reconstructed cryo-EM map (b) and overall structure (c) of the open-spiral pentamer of wild-type MtaLon incubated with ADP. The tensegrity helix triangle (THT) is indicated by the dashed triangle. Map contour levels in ChimeraX: NGDs of the Ob protomers, 0.1; NGDs of the Ac protomers, 0.027; LHs of the Ob protomer, 0.1; LHs of the Ac protomer, 0.05; AAA-protease domains, 0.15. d Arrangement diagram (left) and structures of the protomers in the pentamer, aligned based on the protease domain. The red dash lines are shown to connect the Cα atom of Y397 of Ob1 and other protomers, shown in spheres. A black horizontal line is shown for comparison. e, f Reconstructed cryo-EM map (e) and overall structure (f) of the auto-inhibited spiral hexamer in cylindrical models. Map contour levels: NGDs of the Ob protomers, 0.07; NGDs of the Ac protomers, 0.02; LHs of the Ob protomers, 0.07; LHs of the Ac protomers, 0.01; AAA-protease domains, 0.13. g Arrangement and structures of the protomers in the auto-inhibited hexamer. h, i Reconstructed cryo-EM map (h) and overall structure (i) of the close-ring hexamer of MtaLon-S678A:casein-ADP. Map contour levels: NGDs of the Ob protomers, 0.03; NGDs of the Ac protomers, 0.01; LHs of the Ob protomer, 0.06; LHs of the Ac protomer, 0.05; the substrate and AAA-protease domains, 0.08. j Arrangement and structures of the protomers in the activated hexamer. The red dashed lines are shown to connect the Cα atom of Y397 of Ob1 and other protomers.