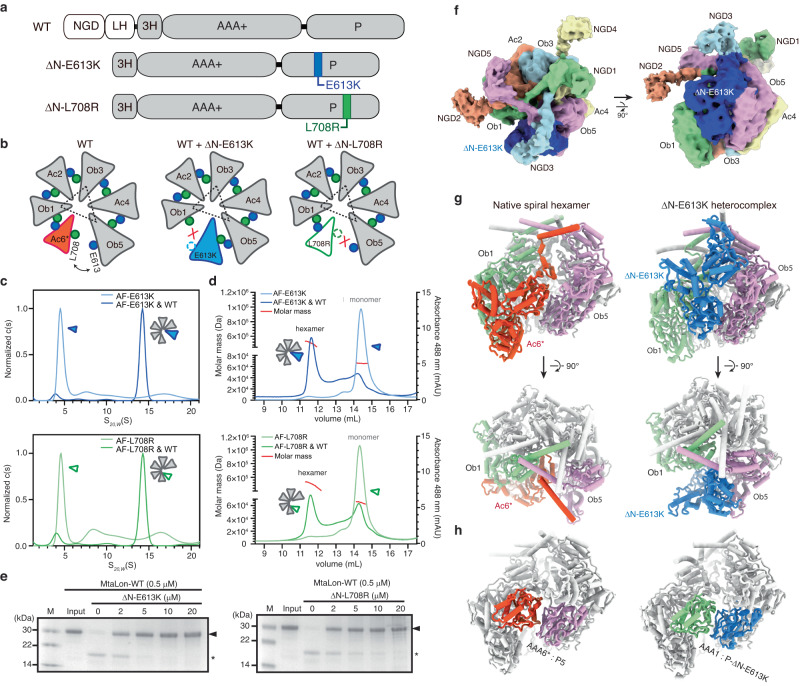
Fig. 4. Designed monomeric mutants bind to the pentameric form and inhibit the activity of Lon.
a Domain diagrams of the mutants ΔN-E613K and ΔN-L708R. b Cartoon illustrations showing the presumed binding modes of ΔN-E613K and ΔN-L708R to the pentameric MtaLon. c SV-AUC analysis of Alexa Fluor 488-labeled ΔN-E613K (AF-E613K), ΔN-L708R (AF-L708R), and their respective incubations with MtaLon (WT). d SEC-MALS analysis of the recycled SV-AUC samples. e Degradation of casein by MtaLon without or in the presence of the indicated amounts of ΔN-E613K and ΔN-L708R. M denotes the molecular weight marker. The “input” lane shows the substrate before the reaction. The substrate α-casein bands in the gels are indicated by the arrowheads, and the asterisks denote product fragments. The experiment was repeated three times independently with similar results. f, g Cryo-EM map (f) and the structure (g) of the heteromeric complex of ΔN-E613K bound to the native pentamer. The structure of the native spiral hexamer is also shown for comparison. Map contour levels in ChimeraX: NGDs of the Ac protomers, 0.036; NGDs of the Ac protomers and AAA-protease domains, 0.1. h Intermolecular interaction between the AAA+ and protease (P) domains, highlighted by the color scheme of (g), in the native spiral hexamer (left) and the ΔN-E613K heterocomplex (right). The NGDs are omitted from the cylindrical models for clarity. Source data are provided as a Source Data file.