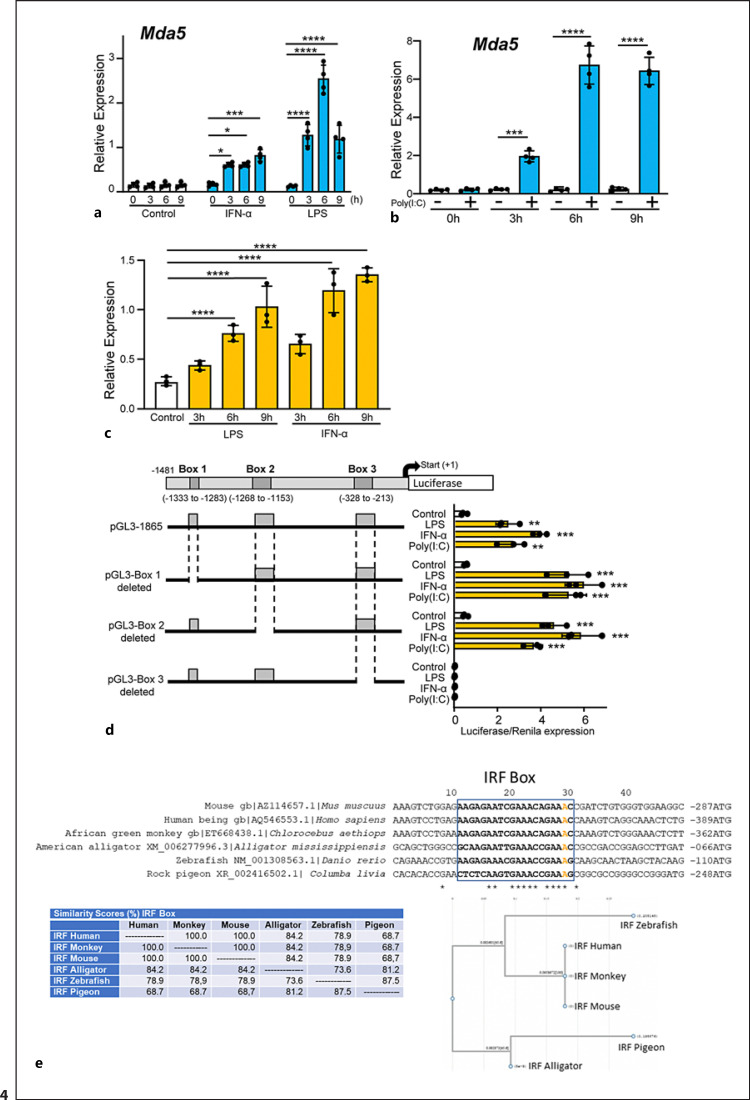
Fig. 4.
Characterization of the Mda5 functional promoter. a RAW 264.7 macrophages were incubated for different times with IFN-α or LPS before Mda5 expression was determined (n = 4). b Similar experiment to that in (a), but using electroporated poly(I:C) as the activator (n = 4). c RAW 264.7 macrophages were transfected with the pGL3−1481 vector. At 24 h post-transfection, cells were stimulated with LPS or IFN-α for the indicated times before luciferase activity was measured. Controls were nonactivated cells (n = 4). d Putative consensus binding sites for transcription factors in the Mda5 gene promoter. The 1481-bp sequence upstream of the Mda5 transcription start site was analyzed using the JASPAR database. Putative boxes showing more than 85% similarity with the consensus sequence for transcription factor binding sites are shown (boxes 1, 2, and 3). As in (c), the vector used contained the indicated deletions of the Mda5 promoter region. Fold increase in Mda5 induction by LPS, IFN-α, or poly(I:C) are shown in relation to the untreated cells (Control). Cells were stimulated for 6h. A plasmid encoding the Renilla luciferase under the control of an HSV-TK promoter was co-transfected, with the Renilla luciferase activity used as a control of transfection. Assays are representative of at least three independent experiments showing similar results. Each experiment was performed in triplicate, and the results are shown as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, and p < 0.0001 in relation to each stimulated sample compared to controls in each experiment. Data were analyzed using the unpaired Student's t test. e Sequences alignment of the Irf1 box in the promoters of Mda5 in human, mice, monkey, alligator, zebrafish and pigeon. JASPAR and the National center for biotechnology information (NCBI) database were used. For sequences alignment we used the MacVector 18.0. The A in orange indicated the distance to the ATG. In the lower part, the similarity (%) between theses boxes in different species is shown, as well as the phylogram.