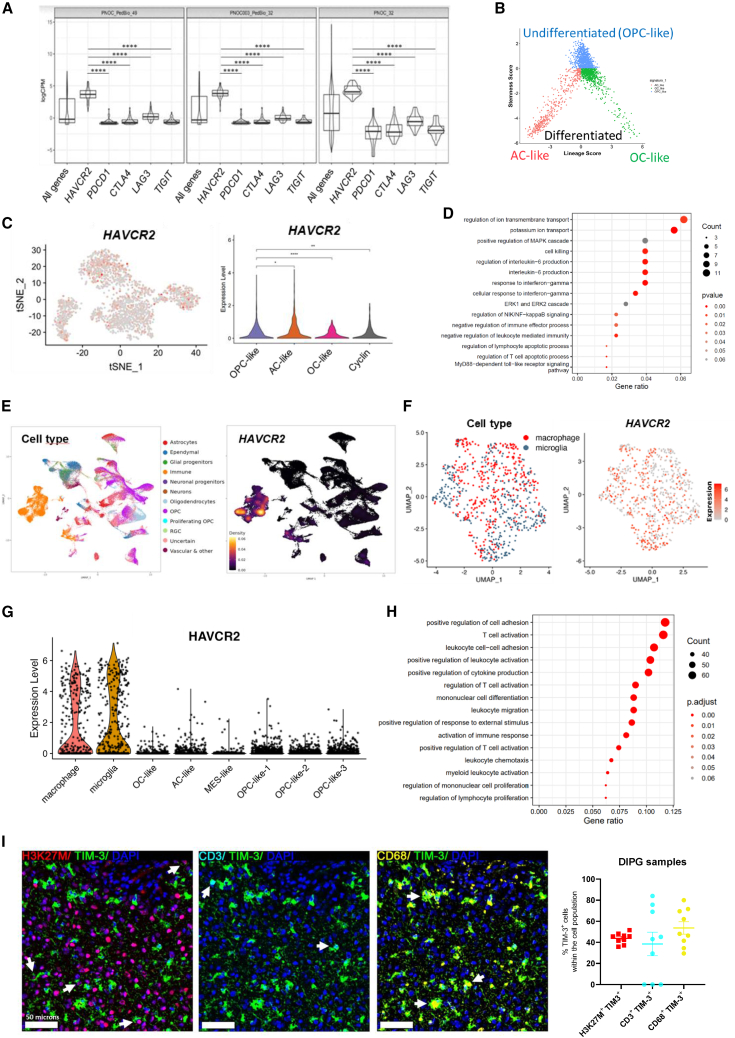
Figure 1.
Evaluation of TIM-3 (HAVCR2) expression in samples from DIPG patients
(A) Violin plots showing the relative mRNA expression of HAVCR2 (TIM-3), PDCD1 (PD-1), CTLA-4, LAG3, and TIGIT in DIPG patients (n = 113) from the PedcBioPortal (PNOC_PedBio_49 and PNOC003_PedBio_32) and Kids First Data Resource Portal (PNOC_32). The lower and upper hinges of the violin plots correspond to the first and third quartiles. After stat (middle): median, 50% quantile, as well as the kernel density estimates as the width.
(B) Two-dimensional representations of the OC-like versus AC-like (x axis) and OPC-like (y axis) scores for DIPG patients.
(C) Left panel, tSNE map of TIM-3 expression in OPC-like, AC-like and OC-like cells. Right panel, Violin plot of TIM-3 expression.
(D) Gene pathways enriched in the set of genes correlated with TIM-3 in OPC-like cells (tumor cells).
(E) Left panel, UMAP plot depicting the different cell populations found in scRNA-seq data for 66 DIPG patients. Right panel, Visualization of HAVCR2 gene expression density in UMAP.
(F) UMAP plot of TIM-3 expression in microglia and macrophages from scRNA-seq data.
(G) Violin plot of TIM-3 expression in microglia, macrophages, OC-like cells, AC-like cells, MES-like cells and OPC-like cells. (H) Gene pathways enriched in the set of genes correlated with TIM-3 in microglia. Data obtained from a previous patient’s scRNA-seq dataset (GSE18435725).
(I) Left panel, representative image from multiplex IF analyses with TIM-3, H3K27M (tumor cells), CD68 (macrophages and microglia) and CD3 (T cells) of DIPG tumor samples from a patient. Right panel, quantification of H3K27M+TIM-3+, H3K27M+TIM-3+, and H3K27M+TIM-3+ compared to total cells in 9 different DIPG patients. One-way ANOVA was performed in panels A and C. Bar graphs indicate the mean ± SEM. (ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). See also Figure S1.