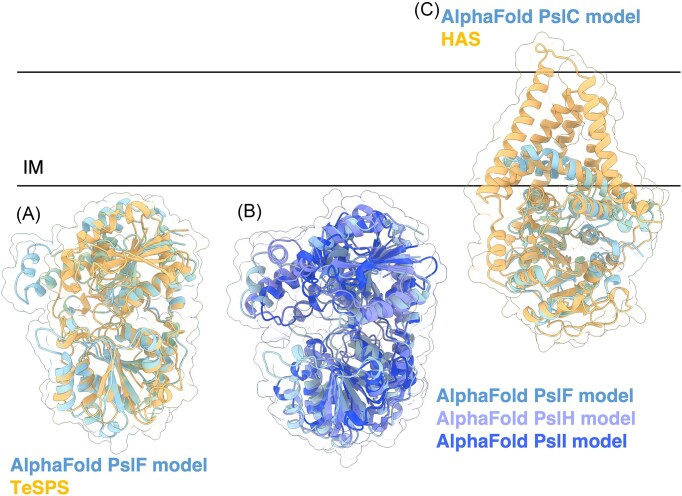
Figure 10.
Structures and predicted AlphaFold models of P. aeruginosa Psl biosynthesis proteins. (A) Superimposition of the P. aeruginosa PslF AlphaFold model (blue) with crystal structure of TeSPS from T. elongatus (orange) (PDB: 6KIH) with a Cα RMSD of 2.54 Å. (B) Superimposition of the P. aeruginosa PslF (blue), PslH (periwinkle), and PslI (dark blue) AlphaFold models. Superimposition of PslH and PslI with PslF generates a Cα RMSD of 3.618 Å and 3.290 Å, respectively. (C) Superimposition of the P. aeruginosa PslC AlphaFold model (blue) with the cryoEM structure of HAS from P. bursaria Chlorella virus CZ-2 (orange) (PDB: 7SP8) with Cα RMSD of 2.836 Å. IM, inner membrane.