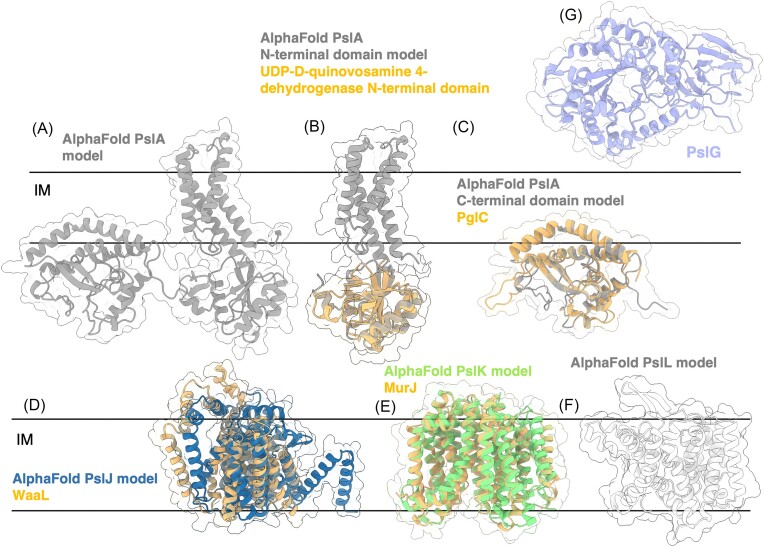
Figure 11.
Structures and predicted AlphaFold models of P. aeruginosa Psl biosynthesis proteins, continued. (A) P. aeruginosa PslA AlphaFold model. (B) Superimposition of the P. aeruginosa PslA AlphaFold model (grey) with the crystal structure of the N-terminal domain of UDP-d-quinovosamine 4-dehydrogenase from V. fischeri (orange) (PDB: 3NKL) with a CαRMSD of 1.337 Å. (C) Superimposition of the C-terminus of P. aeruginosa PslA AlphaFold model (grey) with the crystal structure of the phosphoglycosyl transferase PglC from C. concisus (orange) (PDB: 5W7L) with a Cα RMSD of 1.603 Å. IM, inner membrane. (D) Superimposition of the P. aeruginosa PslJ AlphaFold model (navy) with the cryoEM structure of O-antigen ligase WaaL from Cupriavidus metallidurans (orange) (PDB: 7TPG) with a Cα RMSD of 4.42 Å. (D) Superimposition of the P. aeruginosa PslK AlphaFold model (green) with the crystal structure of the lipid II flippase MurJ from Thermosipho africanus (orange) (PDB: 6NC6) with a Cα RMSD of 2.04 Å. (F) AlphaFold model of P. aeruginosa PslL. (G) Crystal structure of P. aeruginosa PslG (PDB: 5BXA). IM, inner membrane.