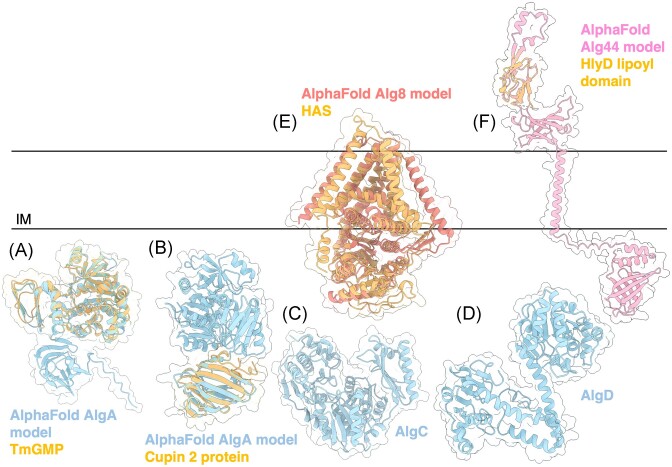
Figure 3.
Structures and predicted AlphaFold models of P. aeruginosa alginate biosynthesis proteins. (A) Superimposition of P. aeruginosa AlgA AlphaFold model (light blue) and the crystal structure of T. maritima GMP TmGMP monomer (orange) (PDB: 2 × 65) with a Cα RMSD of 1.179 Å. (B) Superimposition of P. aeruginosa AlgA AlphaFold model (light blue) and the crystal structure of Shewanella frigidimarina Cupin 2 conserved barrel domain protein (orange) (PDB: 2PFW) with a Cα RMSD of 0.843 Å. (C) Crystal structure of P. aeruginosa AlgC (PDB: 1K2Y). (D) Crystal structure of a monomer of P. aeruginosa AlgD (PDB: 1MV8). (E) Superimposition of P. aeruginosa Alg8 AlphaFold model (salmon) with the crystal structure of the hyaluronan synthase (HAS) from Paramecium bursaria Chlorella virus CZ-2 (orange) (PDB: 7SP8) with a Cα RMSD of 2.932 Å. (F) Superimposition of a monomer of P. aeruginosa Alg44 AlphaFold model (pink) with the crystal structure of the lipoyl domain from the membrane fusion protein HlyD from E. coli (orange) (PDB: 5C22) with a Cα RMSD of 0.774 Å. IM; inner membrane.