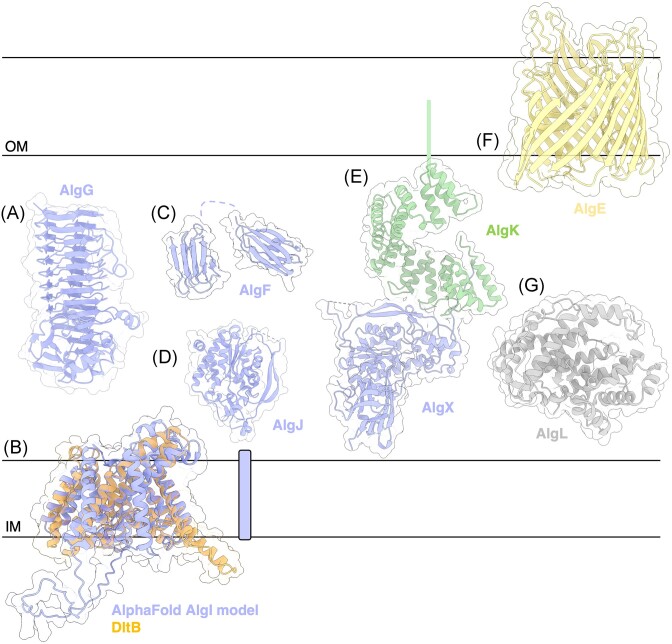
Figure 4.
Structures and predicted AlphaFold models of P. aeruginosa alginate modification and export proteins. (A) Crystal structure of P. syringae pv. Tomato AlgG (PDB: 4NK6). (B) Superimposition of P. aeruginosa AlgI AlphaFold model (periwinkle) and the crystal structure of Streptococcus thermophilus O-acyltransferase DltB (orange) (PDB: 6BUG) with a Cα RMSD of 1.788 Å. (C) CS-Rosetta determined structure of P. aeruginosa AlgF (N-terminus PDB: 6CZT; C-terminus: 6D10). (D) Crystal structure of P. aeruginosa AlgJ (PDB: 4O8V). (E) Crystal structure of P. aeruginosa AlgL (4OZV). (F) Crystal structure of the P. putida AlgKX complex (PDB: 7ULA). (G) Crystal structure of P. aeruginosa AlgE (PDB: 3RBH). (H) Crystal structure of P. aeruginosa AlgL (PDB: 4OZV). IM, inner membrane; OM, outer membrane.