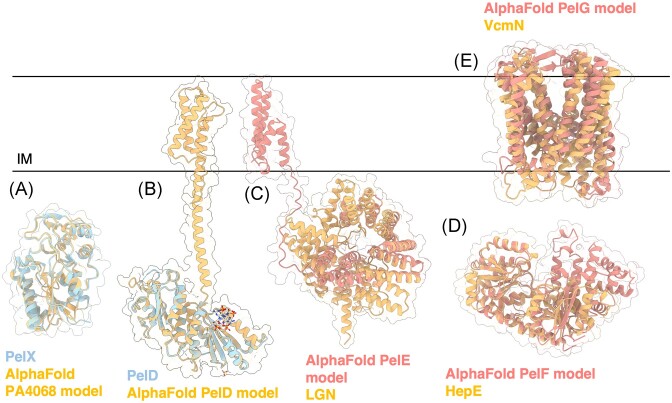
Figure 6.
Structures and predicted AlphaFold models of P. aeruginosa Pel biosynthesis proteins. (A) Superimposition of P. aeruginosa PA4068 AlphaFold model (blue) with the crystal structure of PelX from P. protogens (orange) (PDB: 6WJB) with a Ca RMSD of 0.345 Å. (B) Crystal structure of a monomer of P. aeruginosa PelD (blue) bound to c-di-GMP (PDB: 4DN0) superimposed with the AlphaFold model of PelD (orange). (C) Superimposition of P. aeruginosa PelE AlphaFold model (salmon) with the crystal structure of LGN from Mus musculus (orange) (PDB: 4JHR) with a Ca RMSD of 4.023 Å. (D) Superimposition of P. aeruginosa PelF AlphaFold model (salmon) with the crystal structure of HepE from Anabaena sp. (orange) (PDB: 4XSO) with a Ca RMSD of 1.251 Å. (E) Superimposition of P. aeruginosa PelG AlphaFold model (salmon) with the crystal structure of VcmN from Vibrio cholerae (orange) (PDB: 6IDS) with a Ca RMSD of 5.077 Å. IM, inner membrane.