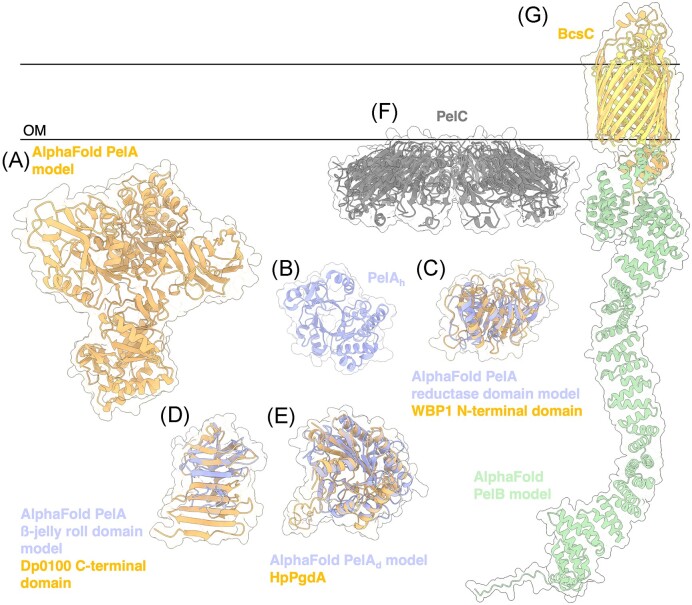
Figure 7.
Structures and predicted AlphaFold models of P. aeruginosa Pel modification and export proteins. (A) AlphaFold model of P. aeruginosa PelA. (B) Crystal structure of the P. aeruginosa PelA hydrolase domain (PelAh) (PDB: 5TCB). (C) Superimposition of P. aeruginosa PelA reductase domain AlphaFold model (periwinkle) with the CryoEM structure of WBP1 from Saccharomyces cerevisiae (orange) (PDB: 6EZN) with a Cα RMSD of 4.650 Å. (D) Superimposition of P. aeruginosa PelA β-jelly roll AlphaFold model (periwinkle) with the crystal structure of the C-terminal domain of Dp0100 alginate lyase from Defluviitalea phaphyphila (orange) (PDB: 6JP4) with a Cα RMSD of 4.379 Å. (E) Superimposition of P. aeruginosa PelA deacetylase (PelAd) domain AlphaFold model (periwinkle) with the crystal structure of HpPgdA (PDB: 4LY4) peptidoglycan deacetylase from Helicobacter pylori (orange) with a Cα RMSD of 3.100 Å. F, Crystal structure of Paraburkholderia phytofirmans PelC (PDB: 5T10). G, Superimposition of P. aeruginosa PelB AlphaFold model (yellow and green) with the crystal structure of BcsC (orange) (PDB: 6TZK) from E. coli with a Cα RMSD of 3.119 Å. OM, outer membrane.