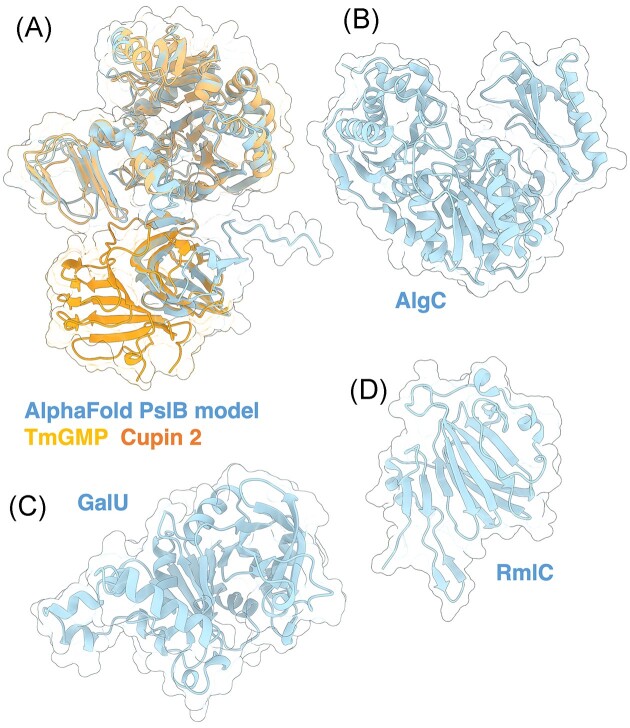
Figure 9.
Structures and predicted AlphaFold models of P. aeruginosa enzymes involved in Psl precursor formation. (A) Superimposition of the P. aeruginosa PslB AlphaFold model (blue) with the crystal structure of T. maritima GMP TmGMP monomer (orange) (PDB: 2 × 65) with a Cα RMSD of 1.403 Å and the crystal structure of S. frigidimarina Cupin 2 conserved barrel domain protein (dark orange) (PDB: 2PFW) with a Cα RMSD of 0.801 Å. (B) Crystal structure of P. aeruginosa AlgC (PDB: 2FKF). (C) P. aeruginosa GalU AlphaFold model. (D) Crystal structure of a monomer of P. aeruginosa RmlC (PDB: 2IXH).