Abstract
Natural genetic variation has facilitated the identification of genes underlying complex traits such as stress tolerances. We here evaluated the long-term (L-) heat tolerance (37°C for 5 days) of 174 Arabidopsis thaliana accessions and short-term (S-) heat tolerance (42°C, 50 min) of 88 accessions and found extensive variation, respectively. Interestingly, L-heat–tolerant accessions are not necessarily S-heat tolerant, suggesting that the tolerance mechanisms are different. To elucidate the mechanisms underlying the variation, we performed a chromosomal mapping using the F2 progeny of a cross between Ms-0 (a hypersensitive accession) and Col-0 (a tolerant accession) and found a single locus responsible for the difference in L-heat tolerance between them, which we named Long-term Heat Tolerance 1 (LHT1). LHT1 is identical to MAC7, which encodes a putative RNA helicase involved in mRNA splicing as a component of the MOS4 complex. We found one amino acid deletion in LHT1 of Ms-0 that causes a loss of function. Arabidopsis mutants of other core components of the MOS4 complex—mos4-2, cdc5-1, mac3a mac3b, and prl1 prl2—also showed hypersensitivity to L-heat stress, suggesting that the MOS4 complex plays an important role in L-heat stress responses. L-heat stress induced mRNA processing–related genes and compromised alternative splicing. Loss of LHT1 function caused genome-wide detrimental splicing events, which are thought to produce nonfunctional mRNAs that include retained introns under L-heat stress. These findings suggest that maintaining proper alternative splicing under L-heat stress is important in the heat tolerance of A. thaliana.
Keywords: selective splicing, natural variation, Arabidopsis thaliana accessions, abiotic stress tolerance
Significance Statement.
Plants are often exposed not only to short-term (S-) heat stress but also to diurnal long-term (L-) heat stress over several consecutive days. However, it is unclear whether plant responses to both S- and L-heat stresses are common or not. We evaluated L-heat tolerances of 170 Arabidopsis thaliana accessions and S-heat tolerances of 88 accessions and found wide variation, respectively. L-heat–tolerant accessions are not necessarily S-heat tolerant, suggesting that the tolerance mechanisms are different. We identified LHT1 as responsible for the variation in L-heat tolerance between Ms-0 and Col-0. LHT1, identical to MAC7, encodes a putative RNA helicase involved in mRNA splicing. Loss of function of LHT1 causes detrimental splicing events, suggesting that LHT1 contributes to maintaining proper alternative splicing under L-heat stress.
Introduction
Since plants cannot move about, they must adapt quickly to environmental stresses. Plants can be subjected to both short-term (S-) severe heat stress, such as at daytime maximum temperatures, and long-term (L-) heat stress over several consecutive days. Nevertheless, most studies of heat tolerance using Arabidopsis thaliana have focused on evaluating S-heat stress, such as 42°C for 30–60 min. Among plant responses to S-heat stress, the accumulation of heat shock proteins (HSPs) and the detoxification of excessive reactive oxygen species (ROS) are thought to be of particular importance and are highly conserved from bacteria to eukaryotes (1–5). Acquired thermotolerance or heat acclimatization is known to increase plant tolerance to subsequent lethal S-heat stress following a nonlethal high-temperature stress event. Epigenetic regulation to maintain higher HSP expression is considered to be important for this acquired thermotolerance (6–8). There are few reports on the isolation of L-heat stress mutants and the identification of their causal genes (9–13), so the mechanism for L-heat stress tolerance is not fully understood. However, these reports show differences in the mechanisms between S-heat and L-heat tolerances. Defects in Arabidopsis Mitochondrial GrpE 2, encoding a nucleotide exchange factor of the HSP70/DnaK complex, result in high sensitivity to L-heat stress, whereas the S-heat tolerance is comparable to that of the wild type (WT) (12). HSP101 plays an important role in S-heat tolerance, but mutation in this gene does not affect L-heat tolerance (12, 14). In contrast, the Arabidopsis heat intolerant 2 (hit2) mutant has defects in both S- and L-heat tolerances, suggesting the existence of common factors in these tolerance mechanisms (11). Indeed, the Arabidopsis hsf1a/b/d/e quadruple mutant had significantly impaired S- and L-heat tolerances, suggesting that the HSFA1s-mediated heat stress response is also important in the L-heat stress response (15). However, several Arabidopsis mutants such as sensitive to long-term heat (sloh) and hit4 have impaired tolerance without reduced expression of HSFs or HSPs, indicating the existence of an HSF-independent pathway (16–18).
Arabidopsis thaliana is distributed widely across the world, and more than 2000 accessions have been collected (19). This broad geographic distribution encompasses substantial variation in nucleotide sequences, and even subtle differences induce large variation in phenotypic traits such as stress tolerance (20–23). Studies of A. thaliana accessions have provided new insights into genome evolution, differentiation among geographic populations, and selective mechanisms that shape complex trait variation in nature (21, 24, 25). Quantitative trait loci for high-temperature germination and fertility under heat stress have been reported (26, 27), yet the variation in heat tolerance during postgermination growth and the underlying mechanisms remain underexplored.
Here, we evaluated L- and S-heat tolerances among A. thaliana accessions. We found wide variation in both, with different mechanisms. We focused on the difference between the L-heat–tolerant Col-0 and that of the L-heat–sensitive Ms-0 to identify the locus contributing to L-heat tolerance and identified LHT1 as responsible for the variation in L-heat tolerance between Ms-0 and Col-0.
Results
Natural variation in L-heat tolerance among A. thaliana accessions
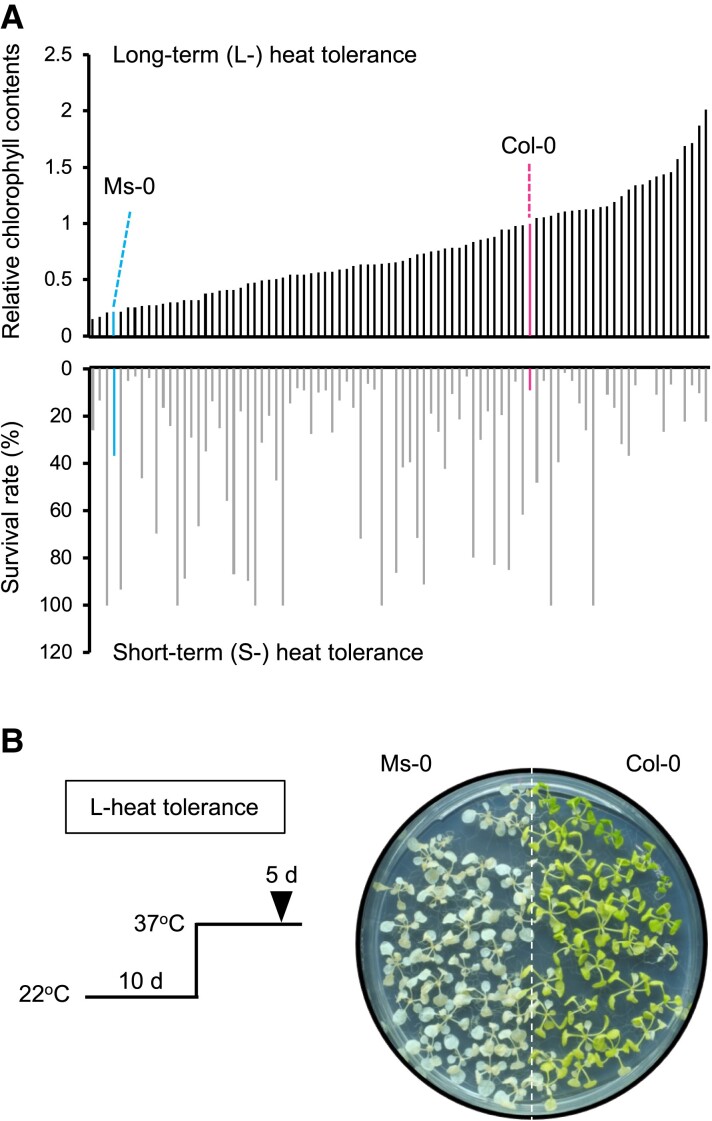
To investigate the natural variation in L-heat tolerance among A. thaliana accessions, we evaluated 170 accessions using the L-heat stress assay (37°C for 5 days) and found nearly a 10-fold variation, represented by the relative chlorophyll content (Fig. 1A, Fig. S1). The reference accession Col-0 had moderate L-heat tolerance. Cvi-0, Ms-0, and Be-1 (as examples) were L-heat sensitive (Fig. 1B). Bs-2, Na-1, Hi-0, and Old-1 were markedly L-heat tolerant (Fig. S2A and B). The L-heat–tolerant accessions were able to survive at 37°C for 7 days, but Col-0 showed complete chlorosis (Fig. S2A). A genome-wide association study (GWAS) using the data of the 170 accessions to identify the genetic loci responsible for the variation found no single-nucleotide polymorphisms (SNPs) that satisfied the genome-wide significance among the 250 k SNP data set (Fig. S3). This result may suggest that L-heat tolerance is a polygenic trait, regulated by multiple loci with relatively small effects, or that the loci contributing to the tolerance differ among accessions. To compare the S- and L-heat tolerances, we evaluated the tolerance of 88 accessions to S-heat stress (42°C for 50 min) and again found ∼10-fold variation in heat tolerance (Fig. 1A, Fig. S4). Ty-0 and Vi-0, among others, proved S-heat tolerant, and Col-0 and Na-1, among many others, proved sensitive (Fig. S4). Interestingly, the L-heat–tolerant accessions were not necessarily S-heat tolerant and vice versa (Fig. 1A, Fig. S5). This result suggests that two traits are governed by different mechanisms.
Fig. 1.
Relationship of L-heat tolerance to S-heat tolerance. A) Comparison of L-heat (37°C for 5 days) and S-heat (42°C for 50 min) tolerances among 88 A. thaliana accessions. Upper panel: L-heat tolerance ranked in order. L-heat tolerance was calculated as chlorophyll content divided by that of Col-0 on the same plate. Lower panel: S-heat tolerance (survival rate after 5 days of S-heat stress) of same accessions as in the upper panel. Accession names are shown in Fig. S5. B) L-heat tolerance of Ms-0 and Col-0.
Identification of Long-term Heat Tolerance 1 locus
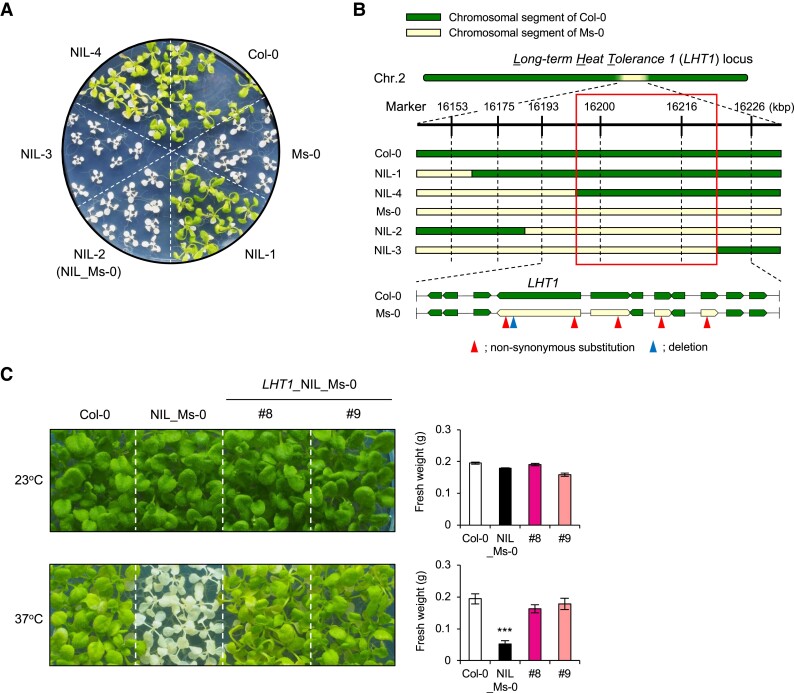
To identify the locus responsible for the variation in L-heat tolerance, we generated F1 progeny of Col-0 and Ms-0, a L-heat–sensitive accession with relatively early flowering. Using F2 progeny, we mapped a single locus on chromosome 2, which we named Long-term Heat Tolerance 1 (LHT1). To narrow down the LHT1 region, we generated near-isogenic lines (NILs) by five rounds of backcrossing of plants showing the L-heat–sensitive phenotype to Col-0 plants. We screened the BC5F2 plants for recombination events within the mapped region containing LHT1 and found NILs of Col-0 and Ms-0 that respectively showed L-heat–tolerant or L-heat–sensitive phenotypes, though they carried a small chromosomal segment from Ms-0 containing the LHT1 region in the genetic background of Col-0 (Fig. 2A and B). To narrow down the LHT1 locus, we determined genotypes within the LHT1 region in NILs #1, #2 (referred to as NIL_Ms-0), #3, and #4 using simple sequence length polymorphism (SSLP) markers. Correlation of these genotypes with L-heat tolerance successfully narrowed the LHT1 locus to 33 kbp (Fig. 2A and B). We next sequenced Ms-0 to detect all polymorphisms at the LHT1 locus between Col-0 and Ms-0. Comparison within the LHT1 region of Col-0 and Ms-0 revealed that of the 11 genes in the region, 3 were polymorphic with nonsynonymous amino acid substitutions and 1 had 2 nonsynonymous substitutions and a 3 bp (1 amino acid) deletion (Fig. 2B). Since F1 progeny between Col-0 and Ms-0 showed L-heat tolerance, we considered that a recessive mutation occurred in Ms-0. Therefore, we cloned the region from promoter to 3′ untranslated region (UTR) of each of four candidate genes from Col-0 and introduced them into L-heat–sensitive NIL_Ms-0. Only the Col-0 gene with a 3 bp deletion in Ms-0 was able to complement the L-heat sensitivity of NIL_Ms-0, suggesting that this is the LHT1 gene responsible for L-heat tolerance (Fig. 2C).
Fig. 2.
Identification of LHT1. A) High-resolution mapping of the LHT1 locus using NILs. L-heat tolerance of Col-0, Ms-0, and NIL-1, 2 (NIL_Ms-0), 3, and 4. B) Graphical genotypes of NILs. The number of the marker represents the position (kbp) on chromosome 2. C) Complementation test for L-heat tolerance using Col-0 type LHT1/MAC7. Ten-day-old seedlings grown at 22°C were placed at 22°C (upper panel) or 37°C (lower panel) for 5 days and then grown at 22°C for 5 days. Right panel: fresh weight of the aerial parts of plants as depicted in the left panel. Differences between Col-0 and NIL_Ms-0 or LHT1_NIL_Ms-0#8 and #9 were analyzed by Student's t test. ***P < 0.001.
Variation in LHT1 gene among A. thaliana accessions
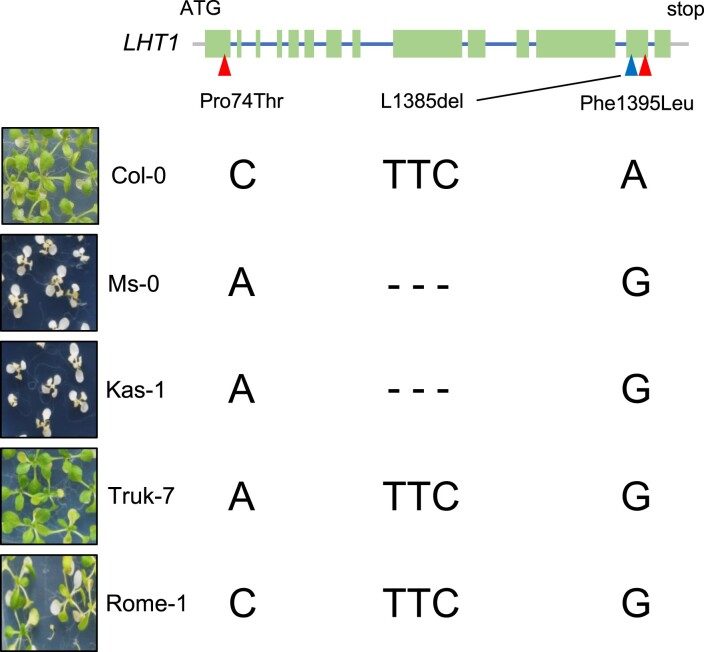
We identified LHT1 by genetic analysis using Col-0 and Ms-0. There were two nonsynonymous substitutions (Pro74Thr and Phe1395Leu) and one amino acid deletion (Leu1385del) in the sequence of LHT1 between Col-0 and Ms-0 (Fig. 3). We examined the phylogenetic tree of A. thaliana accessions based on 143 SNPs and found that Kas-1, Truk-7, and Rome-1 are closely related to Ms-0. To find accessions with an Ms-0-type LHT1 gene considered to have a loss of function, we examined L-heat tolerance and genotypes in Kas-1, Truk-7, and Rome-1. Sequencing revealed that Kas-1 has the same polymorphisms in LHT1 as Ms-0, whereas Truk-7 and Rome-1 had one or two Ms-0-type nonsynonymous substitutions in LHT1 (Fig. 3). Evaluation of L-heat tolerance revealed that only Kas-1, like Ms-0, is sensitive to stress, but Truk-7 and Rome-1 show L-heat tolerance (Fig. 3). These results strongly suggest that one amino acid deletion found in Ms-0 and Kas-1 is responsible for the function of LHT1.
Fig. 3.
Nucleotide diversity of LHT1 gene. Upper panel: Schematic representation of LHT1 gene. Arrowheads indicate polymorphisms with nonsynonymous substitutions (Pro74Thr and Phe1395Leu) and a deletion (L1385del). Left photos: L-heat tolerance of Col-0, Ms-0, Kas-1, Truk-7, and Rome-1. Right nucleotides: genotypes at the Pro74Thr, L1385del, and Phe1395Leu sites.
Functional analyses of LHT1 in L-heat response of A. thaliana
LHT1 is identical to MOS4-ASSOCIATED COMPLEX 7 (MAC7), which encodes an RNA-binding protein with an RNA helicase domain and an intron-binding domain. One amino acid deletion found in LHT1 of Ms-0 was not located in either of these domains but was situated near the RNA helicase domain. T-DNA insertion mutants of LHT1 have an embryonic lethal phenotype, suggesting an important role of LHT1 in plant developmental stages (28). LHT1 has been reported as a component of the MOS4 complex, which is widely conserved in eukaryotes and has been suggested to interact with spliceosomes (29–32). While LHT1 has been suggested to regulate the expression levels of plant disease response genes and to be involved in miRNA biosynthesis and mRNA splicing (33), its involvement in heat or other abiotic stress responses is unknown.
To understand the L-heat stress response of LHT1, we examined the expression profiles under L-heat stress. Induction of LHT1 by L-heat stress suggests that it contributes to the stress response (Fig. 4A). The similar expression pattern in NIL_Ms-0 suggests that the defect in L-heat tolerance in NIL_Ms-0 was not due to reduced expression of LHT1 itself. The fact that the S-heat tolerance of NIL_Ms-0 was comparable to that of Col-0 (Fig. S6) suggests that Ms-0-type LHT1 is specifically defective in mediating L-heat stress response. On the other hand, the better heat tolerance of Ms-0 than of Col-0 suggests that it has loci that contribute to S-heat tolerance independent of LHT1 (Fig. S6).
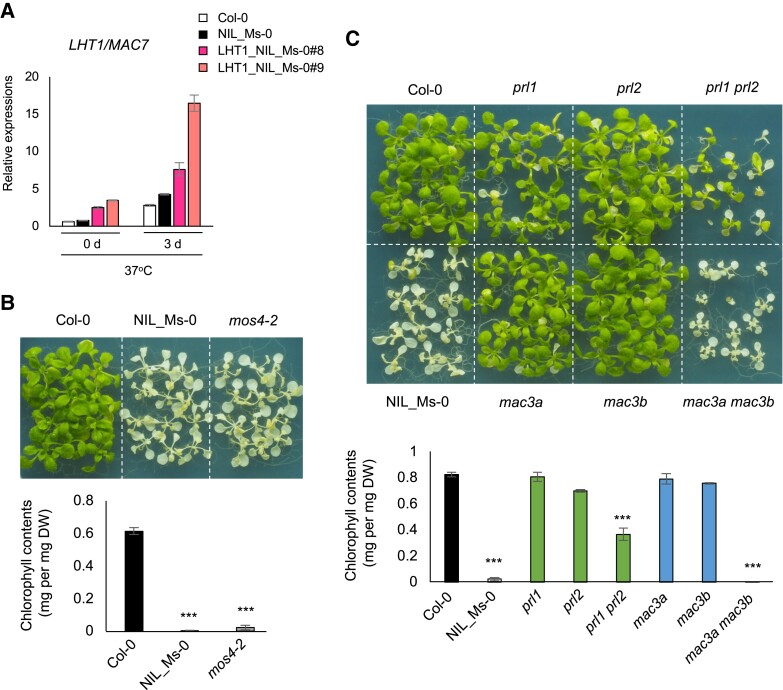
Fig. 4.
L-heat tolerance of mos4 and mutants of MAC genes. A) Expression of LHT1 in Col-0, NIL_Ms-0, and LHT1_NIL_Ms-0#8 and #9 under L-heat conditions, determined by qRT-PCR (mean ± SE, n = 3). B, C) L-heat tolerance of mutants and wild type. Photos show L-heat tolerance of (B) Col-0, NIL_Ms-0, and mos4-2 and (C) prl1, prl2, prl1 prl2, mac3a, mac3b, and mac3a mac3b mutants. Ten-day-old seedlings grown at 22°C were placed at 37°C for 5 days and then grown at 22°C for 5 days. Graphs show chlorophyll contents of plants above. DW, dried weight. Differences between Col-0 wild type and each mutant were analyzed by Student's t test. ***P < 0.001.
We further investigated the contribution of other MOS4 complex components to L-heat tolerance. These factors are known to be involved in plant immune responses, but little is known about abiotic stress responses (33–36). We evaluated the L-heat tolerance of MOS4 core component mutants including MOS4, CDC5, PRL1/2, and MAC3A/B. We generated double mutants for MAC3A and MAC3B and for PRL1 and PRL2 because they have redundant roles in splicing (29, 37). Among them, mos4-2, prl1 + prl2, and mac3a + mac3b had higher sensitivity to L-heat stress than WT (Fig. 4B and C). These results suggest that the MOS4 complex plays an important role in L-heat stress responses.
Effects of Ms-0-type LHT1 on transcriptional regulation
We sampled 10-day-old seedlings of Col-0 and NIL_Ms-0 grown under normal conditions (22°C) and under L-heat stress (37°C for 1, 24, and 72 h) and performed RNA sequencing (RNA-seq) to determine the effect of LHT1 on gene expression. Paired-end sequencing obtained about 60 million reads for each sample. The data were appropriate for analysis because they were clustered within treatment plots and biological repeats by clustering analysis (data not shown).
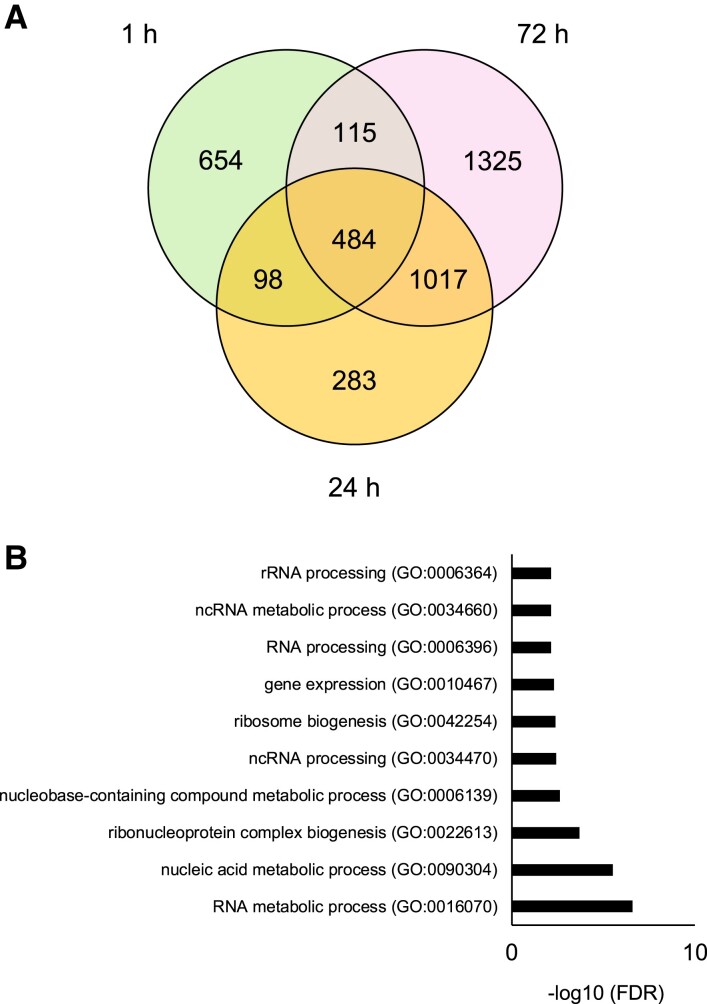
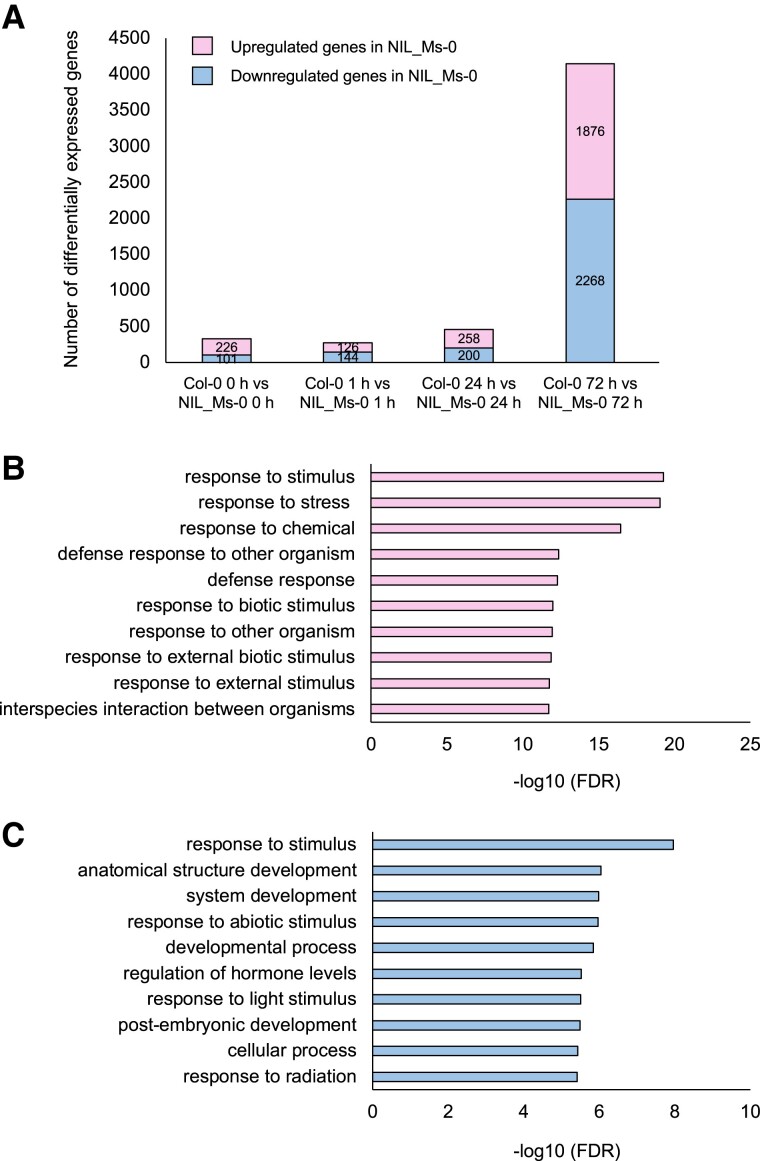
First, we detected heat-induced genes (log2-fold change >2, false discovery rate [FDR] <0.05) of Col-0 at 1, 24, and 72 h (Fig. 5A and Table S1). The group of genes highly expressed at 1 and 24 h included genes that are well known to be induced by heat, such as HSPs and HSFs. On the other hand, the group of genes highly expressed at 72 h included many genes related to RNA processing (Fig. 5B). These results suggest that RNA processing is important for the L-heat stress response. Among genes differentially expressed between Col-0 and NIL_Ms-0 at each time point, the number of genes was significantly higher at 72 h (Fig. 6A and Table S1). Gene Ontology (GO) using Araport software (https://www.araport.org) analysis showed that the up-regulated genes in NIL_Ms-0 at 72 h after heat stress included genes related to defense responses (Fig. 6B and C).
Fig. 5.
Genes induced by L-heat stress in Col-0 WT. A) Venn diagram showing the overlaps between heat-induced genes at 1, 24, and 72 h after heat stress treatment. B) GO analysis of the heat-induced genes specifically at 72 h after heat stress in Col-0. The 1,325 genes were classified according to the GO terms in Araport software into categories based on biological processes.
Fig. 6.
DEGs between Col-0 and NIL_Ms-0 under heat stress. A) Number of DEGs (log2-fold change >2, FDR <0.05) between Col-0 and NIL_Ms-0 at indicated times after heat stress treatment. B, C) GO analysis of the (B) up-regulated and (C) down-regulated genes in NIL_Ms-0 at 72 h after heat stress. The 1,876 genes were classified according to the GO terms in Araport software into categories based on biological processes.
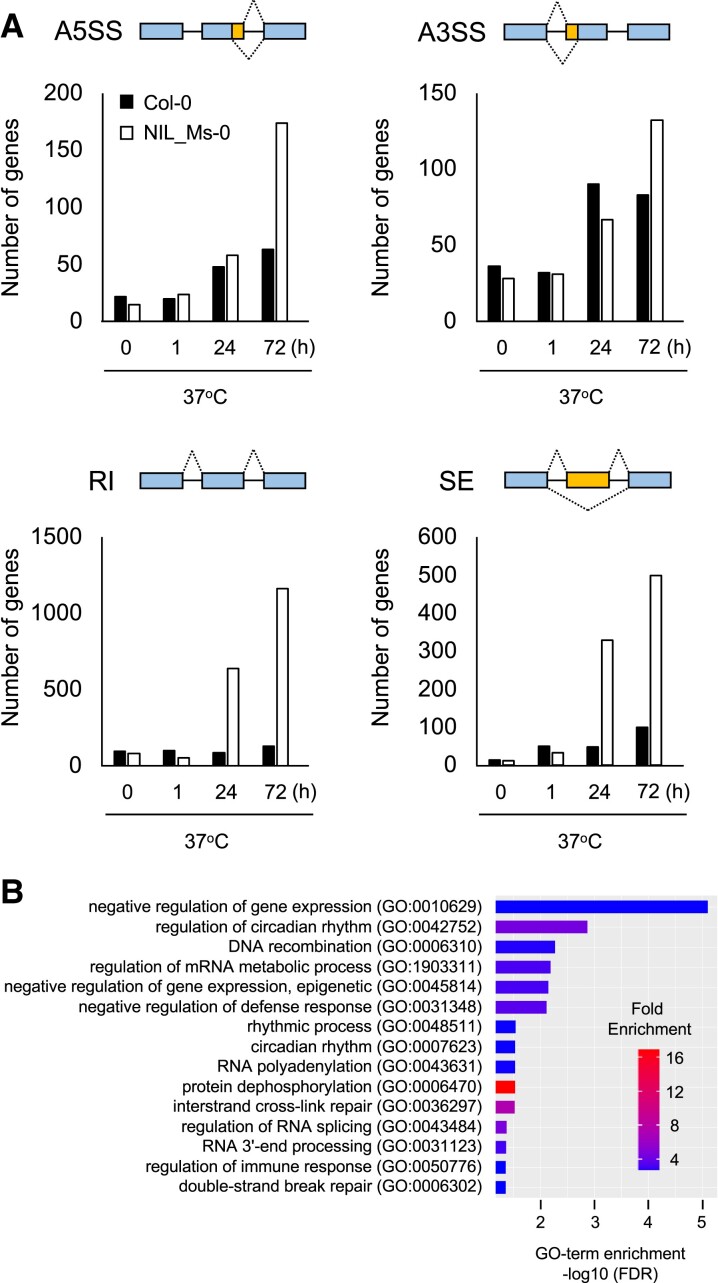
Our findings that LHT1 encodes an RNA-binding protein and that L-heat stress induces RNA processing–related genes implied that the loss of LHT1 function affects alternative splicing under L-heat stress. To test this possibility, we used the RNA-seq data to detect alternative splicing. Alternative splicing can be categorized into four types: exon skipping (SE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), and retained intron (RI) (38). Alternative splicing events in Col-0 and NIL_Ms-0 were compared at 22 and 37°C. At both temperatures at 1 h, there was little difference in the number of splicing events between Col-0 and NIL_Ms-0. During L-heat stress (37°C for 24 and 72 h), however, the number of splicing events increased in NIL_Ms-0, with SE and RI significantly increased (Fig. 7 and Tables S2, S3, and S4). RI is an alternative splicing in which intronic regions are retained, suggesting that increased RI increases nonfunctional mRNAs in NIL_Ms-0 under L-heat stress. These data indicate that all four types of alternative splicing were induced in Col-0 after prolonged heat stress (24 and 72 h; Fig. 7). qRT-PCR to detect the RI of HsfB1 and HSA32 genes in Col-0 and NIL_Ms-0 in order to confirm the validity of the RNA-seq analyses showed that premature RNA levels of both HsfB1 and HSA32 in NIL_Ms-0 under L-heat stress were higher than those in WT (Fig. S7). To characterize the genes showing RI in NIL_Ms-0 at 72 h after heat stress, a GO enrichment analysis was performed. As a result, GO terms including mRNA metabolic process, RNA processing, defense response, and immune response were enriched (Fig. 7B). These GO terms were found as those of genes induced by L-heat stress in Col-0 WT (Fig. 5B) or NIL_Ms-0 (Fig. 6B). It suggests that the genes showing RI in NIL_Ms-0 are consistent with those that were highly expressed under L-heat stress in NIL_Ms-0. It is possible that the effect on splicing caused by one amino acid deletion of LHT1/MAC7 in Ms-0 does not occur in a specific gene but rather in general.
Fig. 7.
Effects on selective splicing between Col-0 and the NIL_Ms-0 under L-heat stress. A) Number of genes showing a specific splicing pattern—selective 5′ splice site (A5SS), selective 3′ splice site (A3SS), RI, and SE under heat stress—at 0, 1, 24, and 48 h. Selective splicing was detected in rMAT software. Genes with a FDR of <0.05 in each comparison were identified as showing selective splicing. B) Applying GO enrichment analysis to the genes showing RI in NIL_Ms-0 at 72 h after heat stress for top 15 biological processes based on positive fold enrichment. The 1,161 genes were classified according to the GO terms in GENEONTOLOGY software (https://geneontology.org/) into categories based on biological processes. The color represents the fold enrichment of the DEGs.
Discussion
Here, we revealed wide variation in both L- and S-heat tolerances of A. thaliana accessions. Interestingly, tolerances to the two stresses were not necessarily related, suggesting that the QTLs responsible for each are different and that the accessions have acquired each stress tolerance independently in their evolution.
Genetic analyses using L-heat–tolerant Col-0 and L-heat–sensitive Ms-0 led us to identify a role of LHT1, a core component of the MOS4 complex, in establishing the L-heat tolerance of A. thaliana. As far as we could determine, the only accession with the Ms-0 genotype with LHT1 was Kas-1, and no GWAS peak was identified at the LHT1 locus, suggesting that this LHT1 genotype is very rare among A. thaliana accessions. The fact that a complete deficiency of LHT1 is embryonic lethal (28) also suggests that there are several accessions in which LHT1 is functionally defective. The Ms-0 genotype with LHT1 is disadvantageous because it impairs L-heat tolerance, but it is unclear what the advantage of having it is. Ms-0 was collected in Moscow, Russia, and Kas-1 was collected in Kashmir, northern India. Since both regions are cold, the disadvantage of the L-heat–sensitive Ms-0 genotype may not have been apparent there.
The absence of SNPs significantly associated with L-heat tolerance in this study suggests that L-heat tolerance is regulated by multiple loci or that the contributing loci differ among accessions. Previously, we isolated several sloh mutants defective in L-heat tolerance, in which the causal genes are different from LHT1 (17, 18). A considerable number of genes may contribute to the variation in L-heat tolerance among A. thaliana accessions. Our large-scale evaluation of L-heat tolerance using A. thaliana accessions revealed that there are several accessions showing more tolerance to L-heat stress than Col-0. The mechanism by which accessions exhibit marked L-heat tolerance is intriguing. Future studies to identify the causal genes in these L-heat–tolerant accessions will lead to further elucidation of the mechanism of L-heat tolerance in A. thaliana. On the other hand, we recently isolated sloh3 and sloh63 mutants, which are hypersensitive to L-heat but not to S-heat stress. SLOH3 and SLOH63 were identical to the splicing-related factor genes MAC9 and MAC17, respectively (Endo et al., in submission as back-to-back article). Interestingly, LHT1/MAC7 interacts with SLOH3/MAC9 and SLOH63/MAC17 (28). Both quantitative trait locus (QTL) analysis and forward genetic screening revealed the importance of maintaining precise mRNA splicing under L-heat stress in Arabidopsis.
QTL analyses have detected loci that contribute to heat tolerance in crops, although the causal gene has not yet been identified. L-heat stress (>35°C for 5 days) at the ear emergence and flowering in rice affects pollen tube elongation and normal pollen dispersal, resulting in the formation of empty or unfertilized grains (39). Thus, most of the recently published heat-tolerant QTLs in rice are related to the ear emergence and flowering stages, and like LHT1, only a few are related to seedling stage QTL (40). A QTL analysis was conducted to identify the loci contributing to heat tolerance in rice at vegetative stage using the recombinant inbred lines obtained by crossing heat-tolerant N22 and heat-sensitive IR64 varieties. A total of 15 loci were detected, of which rlht5.1 was a major QTL contributing 20% toward variation for root length under heat stress (41). In wheat, QTL analysis was also conducted to detect loci affecting wheat yield under drought and heat stress. Of the identified QTL, 17 were related to both stresses, while 16 were related only to heat stress (42). In addition to the fact that no SNPs significantly associated with L-heat tolerance was detected in the GWAS of this study, many QTLs were detected in both wheat and rice, suggesting the complexity of the heat tolerance mechanism in plants. On the other hand, several genes that play a positive role in high-temperature tolerance in rice have been isolated by forward genetics. A heat-sensitive dwarf mutant, thermotolerant growth required1-1, togr1-1, was isolated from an indica variety, Zhongxian 3037. Under L-heat stress (daily maximum temperatures up to 36°C, 15 days exceeding 34°C and 72 days exceeding 30°C), togr1-1 was extremely dwarf with narrow leaf blades and did not set any seed (43). TOGR1 encodes a DEAD-box RNA helicase and functions as a key chaperone for rRNA homeostasis required for rice heat-tolerant growth. It is interesting to note that a different type of RNA helicase than LHT1 contributes to L-heat tolerance in rice.
RNA-seq analyses in this study revealed that L-heat stress induces the expression of genes related to RNA processing. In other studies, transcriptome analysis of Vitis vinifera (European grape) under heat stress conditions suggested that heat stress induces RNA processing–related gene expression, supporting the importance of these genes in the response of plants to heat stress. It has also been reported that deletion of the Arabidopsis splicing factor gene STA1 removes heat acclimatization ability and regulates the splicing of HSF and HSP under heat stress conditions (44). However, HSFs and HSPs were not in the group of genes showing alternative splicing under L-heat stress in this study. These results suggest the existence of a pathway that is important for L-heat response and that is independent of HSP/HSF splicing.
Since LHT1 has been identified as an mRNA-binding protein (28), it may be directly involved in mRNA recognition and regulate L-heat tolerance. Although our results suggest that the precise mRNA splicing regulated by MOS4 complex under L-heat stress is important for stress tolerance, the direct target genes of LHT1 and how LHT1 contributes to the tolerance remain unclear. To elucidate this information, it will be necessary to identify the mRNA to which LHT1 binds and to analyze the function of its splicing variants. It is known that just one splicing variant can critically affect the phenotype (45, 46). Here, NIL_Ms-0, with impaired LHT1 function, affected the splicing of over 3,000 mRNAs. This enormous effect of LHT1 on splicing may affect L-heat tolerance, or there may be a splicing variant that is critical for L-heat tolerance. L-heat stress induced disease-responsive genes in NIL_Ms-0. Mis-activated immunity often results in stunted growth and necrotic lesioning (47). Defects in LHT1 may cause detrimental autoimmunity, thereby reducing L-heat tolerance.
LHT1 is widely conserved in eukaryotes, including in Caenorhabditis elegans as emb-4 (48, 49). EMB-4 binds to introns of target RNAs and is involved in RNA processing and germline gene expression and therefore may also be involved in heat tolerance in C. elegans (48, 49). Indeed, a simultaneous report from our laboratory reveals that the emb-4 mutant and knockdown lines exhibit hypersensitivity to heat stress (Sato et al., in submission as back-to-back article). Taken together, these observations suggest that the heat response mediated by LHT1/emb-4 is evolutionarily conserved between plants and animals.
Materials and methods
Plant materials and growth conditions
We used A. thaliana (L.) Heynh. accessions obtained from the RIKEN BioResource Center, identified by their JA stock number (Table S5). Seeds of the following A. thaliana accessions and mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University): Kas-1(CS28376), Truk-7(CS22805), Rome-1(CS76590), mos4-2 (SALK_090851), cdc5 (SAIL_207_F03), prl1 (SALK_008466), prl2 (SALK_075970), mac3a (SALK_089300), and mac3b (Salk_050811). Seeds were sown on agar (0.8%, w/v) plates (90 mm × 20 mm; Bio-Bik) with 20 mL full-strength Murashige and Skoog (MS) medium containing vitamins (10 mg L−1 myoinositol, 200 µg L−1 glycine, 50 µg L−1 nicotinic acid, 50 µg L−1 pyridoxine hydrochloride, and 10 µg L−1 thiamine hydrochloride, pH 5.7) and 1% sucrose. Plates were sealed with surgical tape. The seeds were stratified at 4°C for 4–7 days and then transferred to a growth chamber (80 µmol photons m2 s−1; 16/8 h light/dark cycle; 22°C) for germination and growth.
Genome-wide association study
GWAS was performed as described (25) to find loci associated with L-heat tolerance in 170 accessions (Table S5).
Plasmid construction and transformation
Plasmids pGreen0029 (50) and pRI909 (Takara, Japan) were used as vectors for plant transformation. DNA fragments from the promoter region to the coding sequence region were amplified using KOD-Plus-Neo (Toyobo, Japan) and introduced into the vector by an In-Fusion HD Cloning Kit (Takara, Japan). pGreen0029 and pRI909 constructs were introduced into Agrobacterium tumefaciens strain GV3101 in the presence of pSoup, a helper plasmid for pGreen replication. Primers for cloning are listed in Table S6. Agrobacteria were then used for plant transformation by the floral dip method. Transgenic plants were selected on MS agar plates containing 200 µg mL−1 claforan and 25 µg mL−1 kanamycin or 20 µg mL−1 hygromycin. Ten-day-old seedlings (T1 plants) were transferred to soil pots.
Heat stress assay
For the L-heat tolerance assay, plates with 10-day-old seedlings were held at 37°C for 5 or 7 days. Aerial parts were harvested and homogenized in cold acetone. To quantify the stress tolerance, chlorophyll content (51) or fresh weight was measured. For the S-heat tolerance assay, plates with 10-day-old seedlings were put into a water bath at 42°C for 40 or 50 min and moved back to 22°C (normal growth conditions) for 5 days. To quantify the stress tolerance, survival rates were measured.
Genetic mapping
The L-heat stress–sensitive Ms-0 was crossed with the tolerant Col-0, and the F1 progeny were self-fertilized to generate F2 populations. Genomic DNA was prepared from individual F2 plants with the recessive L-heat–sensitive phenotype for use as PCR templates. To generate NILs, BC5F2 plants were generated by backcrossing L-heat–sensitive F2 plants (derived from Ms-0 × Col-0) to Col-0 plants five times. We screened the BC5F2 plants for recombination events at the LHT1 locus. Plants were genotyped using SSLP markers listed in Table S6. PCR conditions were consisted of an initial 94°C for 2 min; 30–35 cycles of 94°C for 20 s, 54–57°C for 20 s, and 72°C for 10 s; and a final 72°C for 2 min. The microsatellites were fractionated in 6% agarose gels, and the recombinant value was calculated from the band pattern.
RNA extraction and qRT-PCR
Total RNA was isolated in RNAiso Plus (TaKaRa, Japan) and used as a template to synthesize first-strand cDNA using ReverTra Ace (Toyobo). qRT-PCR was performed on a LightCycler 96 (Roche Diagnostics, Switzerland) with Thunderbird SYBR qPCR Mix (Toyobo) as described (17). ACTIN2 was used an internal standard. Primers are listed in Table S6.
RNA-seq
Total RNA of the extracted samples was quality-checked on an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Libraries were prepared with 500 ng of total RNA obtained from samples with RNA Integrity Number (RIN) >7.0 according to the TruSeq RNA Sample Preparation v. 2 Guide (Illumina, San Diego, CA, USA). Adapters were removed, and quality was filtered in Trimmomatic v. 0.38 software (52), and quality was checked in FastQC v. 0.11.7 software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cleaned reads were mapped to the TAIR10 Arabidopsis genome in HISAT2 (53). The output SAM files were converted to BAM files and sorted in SAMtools v. 1.1 (54). Reads were counted in featureCounts (55) using A. thaliana gene annotation information (http://plants.ensembl.org). Differentially expressed genes (DEGs) were detected in edgeR software (50) from the count data. The BAM files output from the detection of DEGs were used in rMATS software (56) to detect genes with altered selective splicing patterns. The read data were submitted to the DNA Data Bank of Japan (DDBJ) Read Archive (accession number DRA 016172).
GO enrichment analysis
The genes showing RI in NIL_Ms-0 at 72 h after heat stress were applied to GENEONTOLOGY software (https://geneontology.org/) into categories based on biological processes to obtain the GO terms. The obtained GO terms were ordered in descending order for the top 15 biological processes based on positive fold enrichment. These top 15 GO terms are then arranged in descending order of the FDR.
Supplementary Material
Contributor Information
Kazuho Isono, Department of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Kotaro Nakamura, Department of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Kousuke Hanada, Department of Bioscience and Bioinformatics, Kyushu Institute of Technology, Iizuka, Fukuoka 820-8502, Japan.
Kazumasa Shirai, Department of Bioscience and Bioinformatics, Kyushu Institute of Technology, Iizuka, Fukuoka 820-8502, Japan.
Mao Ueki, Department of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Keisuke Tanaka, NODAI Genome Center, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Takashi Tsuchimatsu, Department of Biological Sciences, University of Tokyo, Tokyo 113-0033, Japan.
Satoshi Iuchi, RIKEN BioResource Research Center, Tsukuba, Ibaraki 305-0074, Japan.
Masatomo Kobayashi, RIKEN BioResource Research Center, Tsukuba, Ibaraki 305-0074, Japan.
Izumi Yotsui, Department of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Yoichi Sakata, Department of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Teruaki Taji, Department of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
This work was supported by KAKENHI grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21H05668, 23H04206, and 23H00334 to Te.T.).
Author Contributions
K.I. and Te.T. initiated, conceived, and coordinated the project; K.N. and K.I. evaluated the L-heat tolerance of Arabidopsis; Ta.T. performed GWAS; K.I. generated NILs and identified the LHT1/MAC7 gene responsible for L-heat–sensitive phenotype of Ms-0; K.I. performed physiological analyses; S.I. and M.K. provided seeds of A. thaliana accessions and found a closely related accession of Ms-0; K.I. and K.T. performed RNA-seq and analysis; K.I. and K.H. detected splicing variants; Te.T. wrote the manuscript with assistance from I.Y. and Y.S.
Data Availability
All data that support the findings of this study are available within its supplementary materials.
References
- 1. Liu H-C, Liao H-T, Charng Y-Y. 2011. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34:738–751. [DOI] [PubMed] [Google Scholar]
- 2. Mishra SK, et al. 2002. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16:1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. 2017. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22:53–65. [DOI] [PubMed] [Google Scholar]
- 4. Vanderauwera S, et al. 2011. Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci USA. 108:1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshida T, et al. 2011. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics. 256:321–332. [DOI] [PubMed] [Google Scholar]
- 6. Brzezinka K, et al. 2016. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 5:e17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charng Y-Y, Liu H-C, Liu N-Y, Hsu F, Ko S. 2006. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 140:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamaguchi N, et al. 2021. H3k27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat Commun. 12:3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee C-F, et al. 2006. Mutation in a homolog of yeast Vps53p accounts for the heat and osmotic hypersensitive phenotypes in Arabidopsis hit1-1 mutant. Planta 224:330–338. [DOI] [PubMed] [Google Scholar]
- 10. Wang L-C, et al. 2011. Involvement of the Arabidopsis HIT1/AtVPS53 tethering protein homologue in the acclimation of the plasma membrane to heat stress. J Exp Bot. 62:3609–3620. [DOI] [PubMed] [Google Scholar]
- 11. Wu S-J, Wang L-C, Yeh C-H, Lu C-A, Wu S-J. 2010. Isolation and characterization of the Arabidopsis heat-intolerant 2 (hit2) mutant reveal the essential role of the nuclear export receptor EXPORTIN1A (XPO1A) in plant heat tolerance. New Phytologist 186:833–842. [DOI] [PubMed] [Google Scholar]
- 12. Hu C, Lin S, Chi W, Charng Y. 2012. Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol. 158:747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu J-R, et al. 2016. The Arabidopsis heat-intolerant 5(hit5)/enhanced response to aba 1(era1) mutant reveals the crucial role of protein farnesylation in plant responses to heat stress. New Phytologist 213:1–13. [DOI] [PubMed] [Google Scholar]
- 14. Queitsch C, Hong SW, Vierling E, Lindquist S. 2000. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han S-H, Park Y-J, Park C-M. 2020. HOS1 activates DNA repair systems to enhance plant thermotolerance. Nat Plants. 6:1–20. [DOI] [PubMed] [Google Scholar]
- 16. Wang L-C, et al. 2013. Arabidopsis HIT4 encodes a novel chromocentre-localized protein involved in the heat reactivation of transcriptionally silent loci and is essential for heat tolerance in plants. J Exp Bot. 64:1689–1701. [DOI] [PubMed] [Google Scholar]
- 17. Isono K, et al. 2020. An ER–Golgi tethering factor SLOH4/MIP3 is involved in long-term heat tolerance of Arabidopsis. Plant Cell Physiol. 62:272–279. [DOI] [PubMed] [Google Scholar]
- 18. Tsukimoto R, et al. 2021. Mitochondrial fission complex is required for long-term heat tolerance of Arabidopsis. Plant Cell Physiol. 63:296–304. [DOI] [PubMed] [Google Scholar]
- 19. 1001 Genomes Consortium . 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gehan MA, et al. 2015. Natural variation in the C-repeat binding factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J. 84:682–693. [DOI] [PubMed] [Google Scholar]
- 21. Katori T, et al. 2010. Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J Exp Bot. 61:1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rus A, et al. 2006. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2:e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kobayashi Y, et al. 2008. Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 148:969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mitchell-Olds T, Schmitt J. 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441:947–952. [DOI] [PubMed] [Google Scholar]
- 25. Ariga H, et al. 2017. NLR locus-mediated trade-off between abiotic and biotic stress adaptation in Arabidopsis. Nat Plants. 3:1–8. [DOI] [PubMed] [Google Scholar]
- 26. Bac-Molenaar JA, et al. 2015. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell. 27:1857–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoong F-Y, et al. 2016. Genetic variation for thermotolerance in lettuce seed germination is associated with temperature-sensitive regulation of ETHYLENE RESPONSE FACTOR1 (ERF1). Plant Physiol. 170:472–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jia T, et al. 2017. The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell. 29:2626–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monaghan J, et al. 2009. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5:e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson K, Dong O, Li X. 2011. The evolutionarily conserved MOS4-associated complex. Open Life Sci. 6:776–784. [Google Scholar]
- 31. Koncz C, deJong F, Villacorta N, Szakonyi D, Koncz Z. 2012. The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front Plant Sci. 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng X, et al. 2016. Recruitment of the NineTeen complex to the activated spliceosome requires AtPRMT5. Proc National Acad Sci. 113:5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palma K, et al. 2007. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Gene Dev. 21:1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang S, Xie M, Ren G, Yu B. 2013. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc National Acad Sci. 110:17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang S, Liu Y, Yu B. 2014. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 10:e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li S, et al. 2018. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 30:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weihmann T, Palma K, Nitta Y, Li X. 2012. PLEIOTROPIC REGULATORY LOCUS 2 exhibits unequal genetic redundancy with its homolog PRL1. Plant Cell Physiol. 53:1617–1626. [DOI] [PubMed] [Google Scholar]
- 38. Dikaya V, et al. 2021. Insights into the role of alternative splicing in plant temperature response. J Exp Bot. erab234. [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39. Satake T, Yoshida S. 1978. High temperature-induced sterility in Indica rices at flowering. Jpn J Crop Sci. 47:6–17. [Google Scholar]
- 40. Liu H, et al. 2023. Genetic research progress: heat tolerance in rice. Int J Mol Sci. 24:7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kilasi NL, et al. 2018. Heat stress tolerance in rice (Oryza sativa L.): identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Front Plant Sci. 9:1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pinto RS, et al. 2010. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor Appl Genet. 121:1001–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang D, et al. 2016. Nucleolar DEAD-box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet. 12:e1005844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim G-D, Cho Y-H, Lee B-H, Yoo S-D. 2017. STABILIZED1 modulates pre-mRNA splicing for thermotolerance. Plant Physiol. 173:2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhan X, et al. 2015. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat Commun. 6:8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gu J, et al. 2017. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 46:gkx1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alcázar R, Parker JE. 2011. The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16:666–675. [DOI] [PubMed] [Google Scholar]
- 48. Akay A, et al. 2017. The helicase Aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev Cell. 42:241–255.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tyc KM, et al. 2017. The conserved intron binding protein EMB-4 plays differential roles in germline small RNA pathways of C. elegans. Dev Cell. 42:256–270.e6. [DOI] [PubMed] [Google Scholar]
- 50. Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. 2002. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 42:819–832. [DOI] [PubMed] [Google Scholar]
- 51. Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA)—Bioenergetics. 975:384–394. [Google Scholar]
- 52. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. [DOI] [PubMed] [Google Scholar]
- 56. Shen S, et al. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc National Acad Sci. 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available within its supplementary materials.