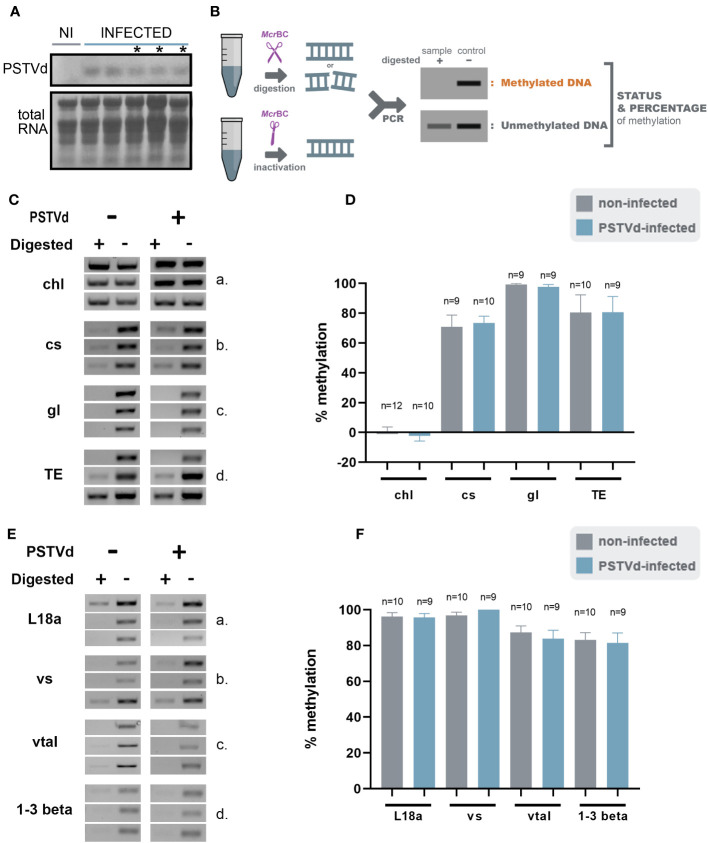
Figure 2.
McrBc digestions in non-infected and PSTVd-infected plants. (A) Northern blot analysis of WT and PSTVd-infected N. benthamiana plants (3 wpi). Total RNA identified with methylene blue was used as a loading control. (*) stands for samples used for further analysis. (B) Schematic representation of the McrBC principle. (C, E) Chop PCRs using MrcBC in DNA of WT and PSTVd-infected plants. Extracted DNA was cut with McrBC followed by PCR amplification. If methylated no band is observed in the digested sample. Enzyme inactivation was used as control. Used targets were a control DNA (chl - C,a), promoter region of cellulose synthase (cs - C,b), germin-like (gl – C,c), TE Cc), ribosomal L18a (E,a), vironine synthase (vs - E,b), vacuolar protein sorting - associated protein (vtaI - E,c) and glucan endo 1-3 beta – glucosidase (E,d). Triplicates were made for each digestion. One is presented here and two more in Figure S2 (D, F) Graphical representation of chop PCRs. ‘n’ represents the number of calculated replicates from all biological and technical repeats.