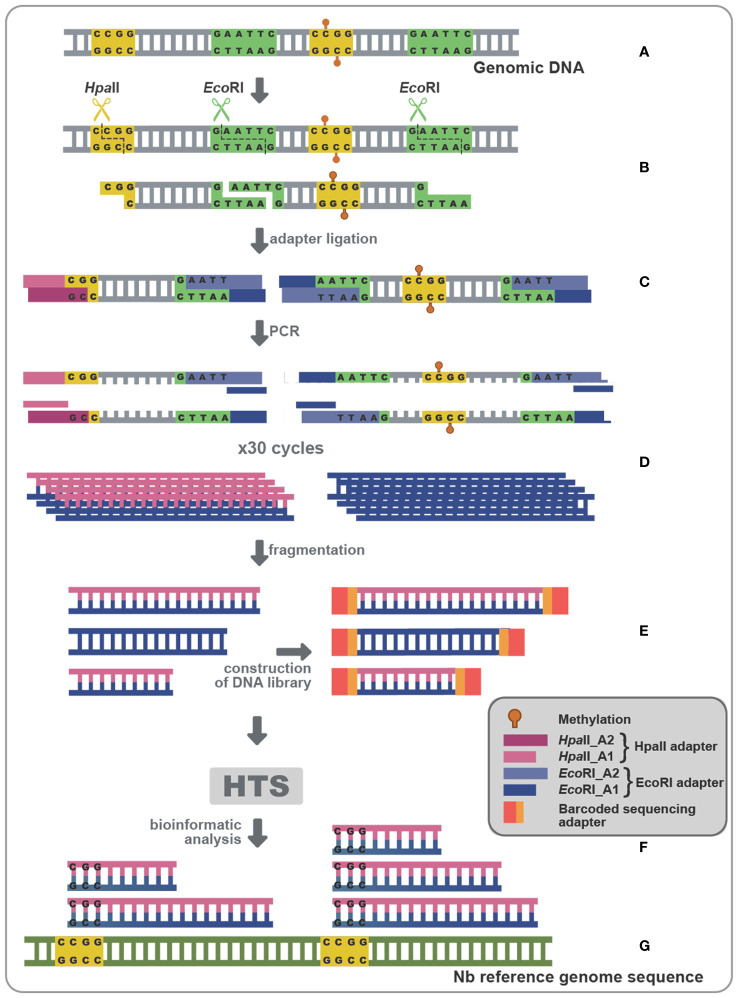
Figure 6.
MSAP-Seq. Schematic representation of the MSAP-Seq pipeline. After genomic extraction with an appropriate protocol (A), EcoRI, a rare cutter and HpaII, a MRSE recognizing unmethylated cytosines in the pattern CCGG were used (B), followed by ligation of adapters (HpaII-A1/HpaII-A2 and EcoRI-A1/EcoRI-A2) containing a known region (C), which allowed for a PCR amplification (D). Then, a sonication step drives fragmentation of PCR parts, followed by a library construction (E) and a High-throughput sequencing (HTS) (F). Finally, results are analyzed using a bioinformatic pipeline (G).