Figure 3.
Systematic predictive performance evaluation of inclusive PGS (iPGS) models and PRS-CSx across 60 anthropometric and hematological traits in the UK Biobank
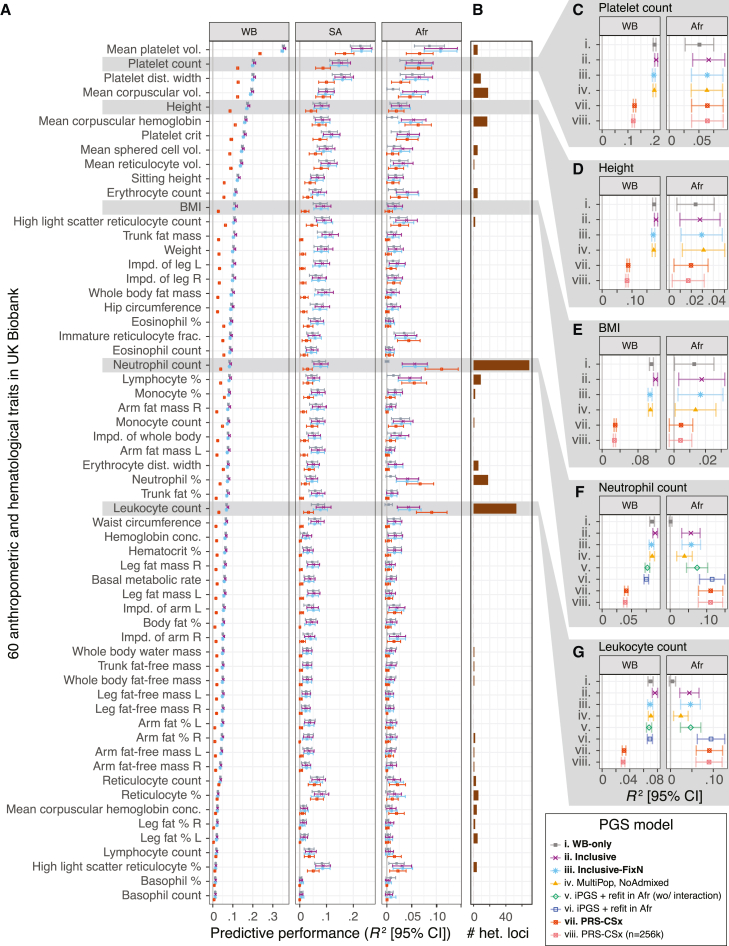
(A) The predictive performance (R2) in White British (WB), South Asian (SA), and African (Afr) groups in the UK Biobank are shown for four select models: (i) WB-only, (ii) inclusive, (iii) inclusive-FixN, and (vii) PRS-CSx.
(B) The number of approximately LD-independent (R2 < 0.2 in the African population in the UK Biobank) variants with heterogeneous GWAS associations (material and methods).
(C–G). The predictive performance of up to eight PGS models in White British (WB) and African (Afr) populations in the UK Biobank are shown for five select traits. The refit models are trained only for the neutrophil and leukocyte counts, where genetic variants with heterogeneous GWAS effects were observed. The predictive performance for other models and ancestry groups is shown in Figures S4 and S5. BMI: body mass index. Vol.: volume. Dist.: distribution. Impd.: impedance. Frac.: fraction. Conc.: concentration. %: percentage. R: right. L: left. Error bars represent 95% confidence intervals.