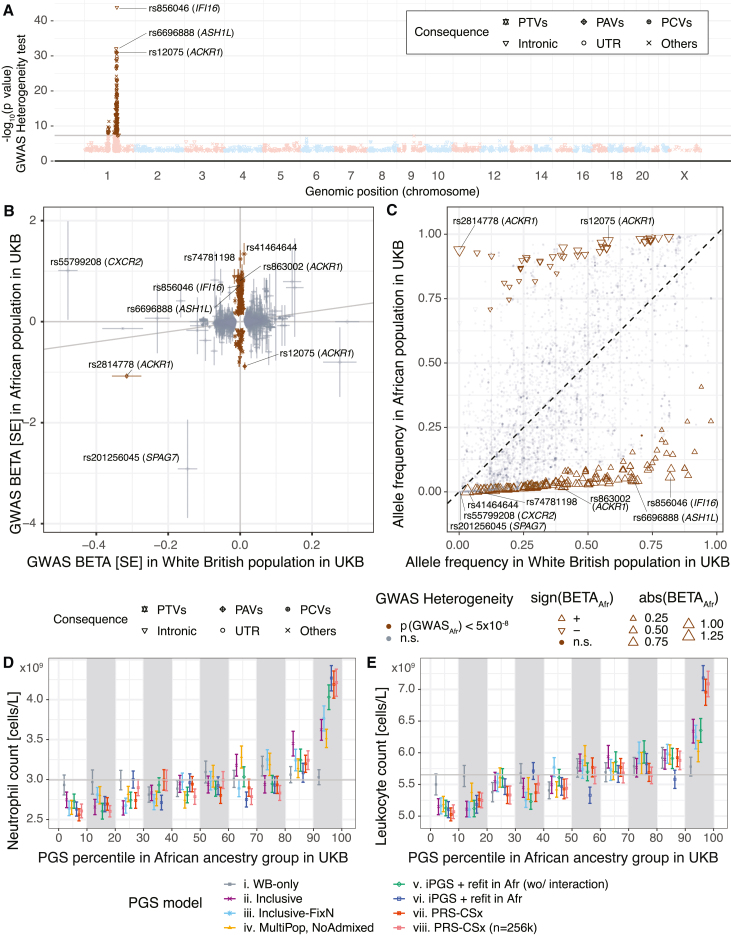
Figure 4.
Enhanced predictive performance with iPGS+refit that additionally accounts for ancestry-dependent genetic effects
(A) GWAS meta-analysis heterogeneity test in the UK Biobank for the neutrophil count. Genetic variants with heterogeneity p value < 5 × 10−8 are highlighted in brown.
(B) GWAS effect size comparison between White British (x axis) and African (y axis) populations in the UK Biobank. The color indicates whether the variants show heterogeneous GWAS associations. Error bars represent the standard error of the GWAS effect-size estimates.
(C) Allele frequency comparison for 5,890 genetic variants associated with neutrophil count (material and methods). The color indicates whether the variants show heterogeneous GWAS associations. The shape and size represent the direction and the magnitude of GWAS associations in the African population in the UK Biobank.
(D and E) Phenotype mean values of neutrophil count (D) and leukocyte count (E) stratified by decile of PGS in the held-out test set of individuals of the African population in the UK Biobank are shown. Error bars represent the standard error-of-mean estimates.
PTVs: protein-truncating variants. PAVs: protein-altering variants. PCVs: proximal coding variants. Intronic: intronic variants. UTR: genetic variants on untranslated regions. Others: other non-coding variants.