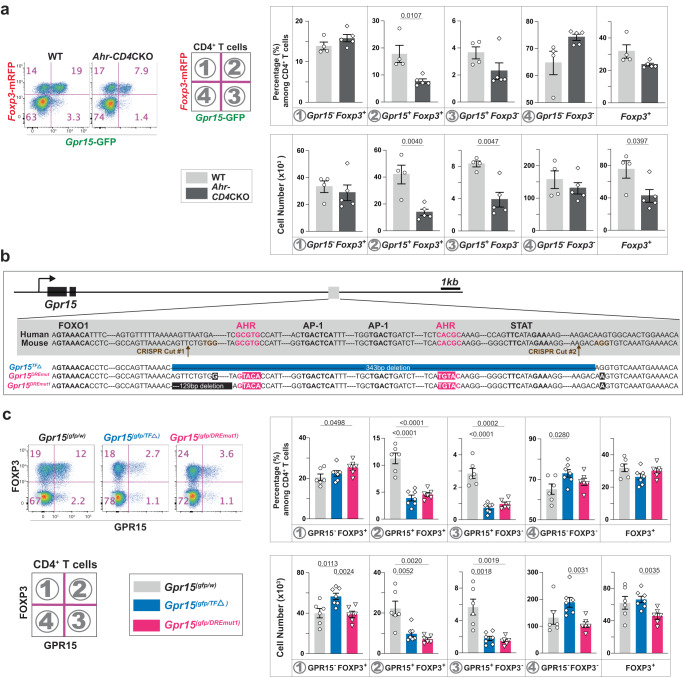
Fig. 5. AhR directly controls the transcriptional activation of Gpr15 in CD4+ T cells via dioxin-responsive elements (DRE).
a Gpr15 and Foxp3 mRNA expression in CD4+ T cells in the LILP was determined by the reporter expression in the presence (WT: Ahr(fl/fl)Gpr15(gfp/+)Foxp3mrfp, n = 4) or the absence (Ahr-CD4CKO: CD4CreAhr(fl/fl)Gpr15(gfp/+)Foxp3mrfp, n = 5) of AhR. Flow cytometry plots and the percentage/cell number of each population among CD4+ T cells in the LILP are shown. Representatives of at least two independent experiments. b The DNA sequence of the conserved region in the 3′-distal to Gpr15 locus. Our knock-in strategy is shown: Gpr15TFΔ allele has a deletion of 343 bps containing two AhR binding sites, two AP-1 binding sites, and a STAT binding site. Also, we intended to mutate two AhR binding sites to make a Gpr15DREmut allele but ended up with a Gpr15DREmut1 allele having an additional 129 bp deletion of the non-conserved sequence. c Representative flow cytometry plots and the percentage/cell number of two independent experiments are shown for CD4+ T cells in the LILP. The number of mice used: 6 for WT, 7 for Gpr15TFΔ, and 6 for a Gpr15DREmut1. 8–14-week-old mice were used (a, c). Data are presented as mean values±SEM (a, c). Each data point represents the result from one mouse, and p values were calculated by two-sided student’s t-test (a, c). Source data are provided as a Source Data file (a, c).