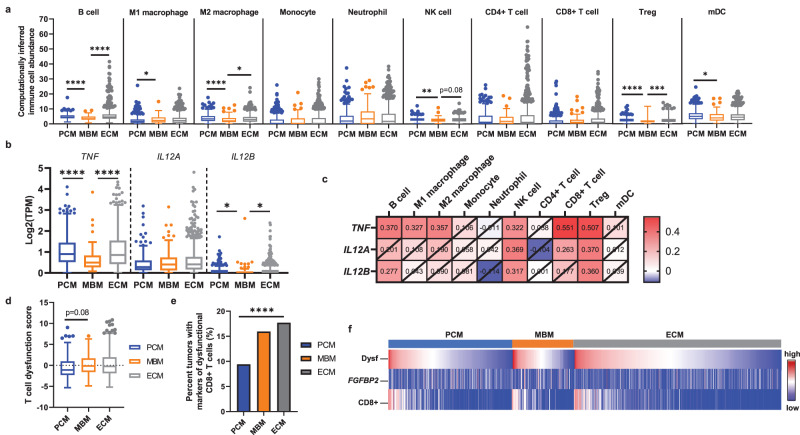
Fig. 2. MBM have significantly fewer computationally inferred immune cell infiltrates and more T cell dysfunction.
a Box plots showing computationally inferred immune cell abundance in PCM, MBM, and ECM using quanTIseq analysis. The data is displayed using the Tukey method for box and whiskers, with the center line indicating the median. Kruskal-Wallis test with Benjamini-Hochberg correction; *, corrected p < 0.05; **, corrected p < 0.005; ***, corrected p < 0.0005; ****, corrected p < 0.0001. b The mRNA levels of TNF, IL12A, and IL12B were compared between PCM, MBM, and ECM tumor samples. The data is displayed using the Tukey method for box and whiskers, with the center line indicating the median. Kruskal–Wallis test with Benjamini-Hochberg correction; **, corrected p < 0.005; ****, corrected p < 0.0001 c Heat map of Spearman rank correlation coefficients between TNF, IL12A, and IL12B mRNA and immune cell infiltrates calculated from bulk transcriptomic data in MBM. The numbers in each box show the correlation coefficient, where crossed boxes indicate non-significant correlation values (p > 0.05). d Box plots of T cell exhaustion scores in PCM, MBM, and ECM. The data is displayed using the Tukey method for box and whiskers. Kruskal–Wallis test with Benjamini–Hochberg correction. e Percentage of PCM, MBM, and ECM with CD8 + T cells that have high dysfunction scores (composite z-score > 1.0) but low FGFBP2 mRNA levels (less than the median for the whole cohort). χ2 test; ****p < 0.0001. f Oncoplot with T cell dysfunction score (top, highest to lowest within PCM, blue; MBM, orange; ECM, gray), FGFBP2 mRNA levels (middle), and CD8 + T cell abundance (bottom).