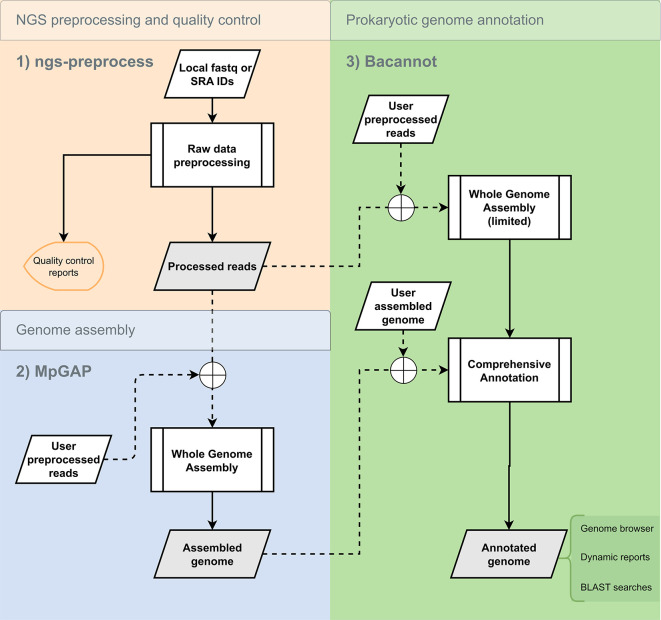
Figure 1. Flowchart of optional sequential execution of the developed pipelines for complete bacterial genomics analysis.
The available whole genome assembly module in bacannot is considered limited compared to MpGAP, because it only contains two assembler options. Dashed arrows connected to the crossed circles represent the optional flow of data, highlighting that chaining the pipelines is not required. Gray boxes highlight the pipeline’s outputs.