Figure 4. Secondary structures are not pre‐formed and likely arise as a result of replication, leading to helicase‐polymerase uncoupling.
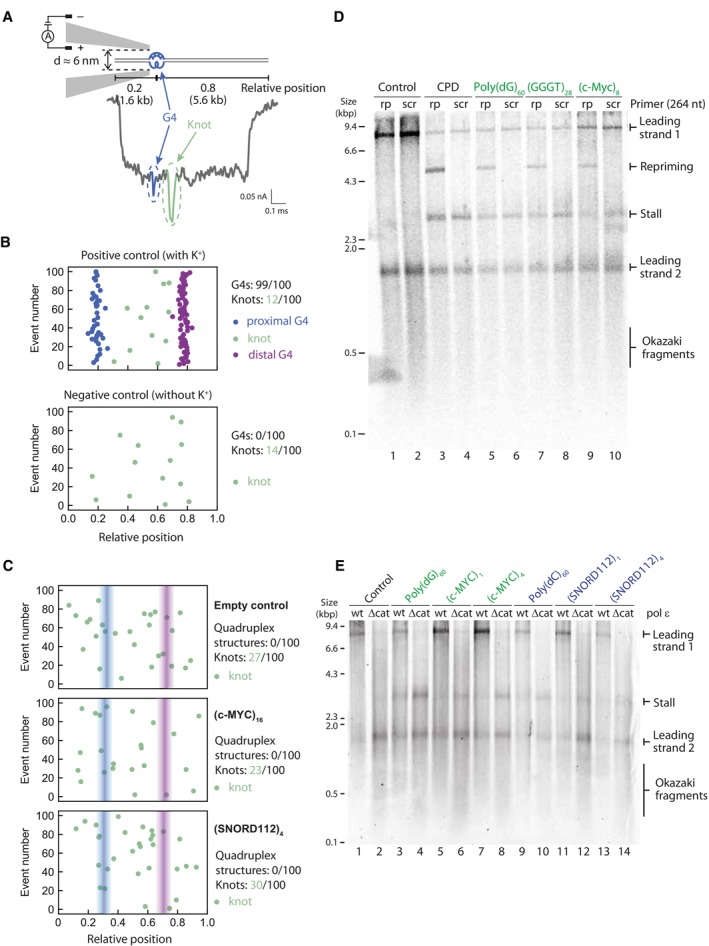
- Schematic of a positive control DNA passing through the nanopore in the direction that positions the G4 proximally and a representative nanopore measurement event with a DNA knot in the middle of the molecule. The G‐quadruplex structure and its corresponding current drop are marked in blue. The DNA knot and its corresponding current drop are marked in green. Numbers indicate the proportion through the DNA the G4 is positioned.
- Summary scatterplots of the peak positions in the first 100 informative nanopore events for the positive control (where K+ is added) and the negative control (no K+ present). The negative control does not contain any G‐quadruplex structure without the presence of potassium ions. Numbers indicate proportion of G4s or knots in 100 unfolded events.
- Nanopore measurement results of the first 100 informative nanopore events for replication templates. Summary scatterplots of the peak positions are shown for an empty control and substrates containing (c‐MYC Pu22)16 or (SNORD112)4. Numbers indicate proportion of G4s or knots in 100 informative events.
- Replication reactions carried out on G4‐containing templates in the presence of a primer that anneals 264 nt downstream of the G4 (rp) or a scrambled control primer (scr).
- Replication of G4 or iM substrates with either wildtype pol ε (wt) or a pol ε mutant with a deleted catalytic domain (Δcat). Products were analysed on a denaturing agarose gel.
Source data are available online for this figure.
