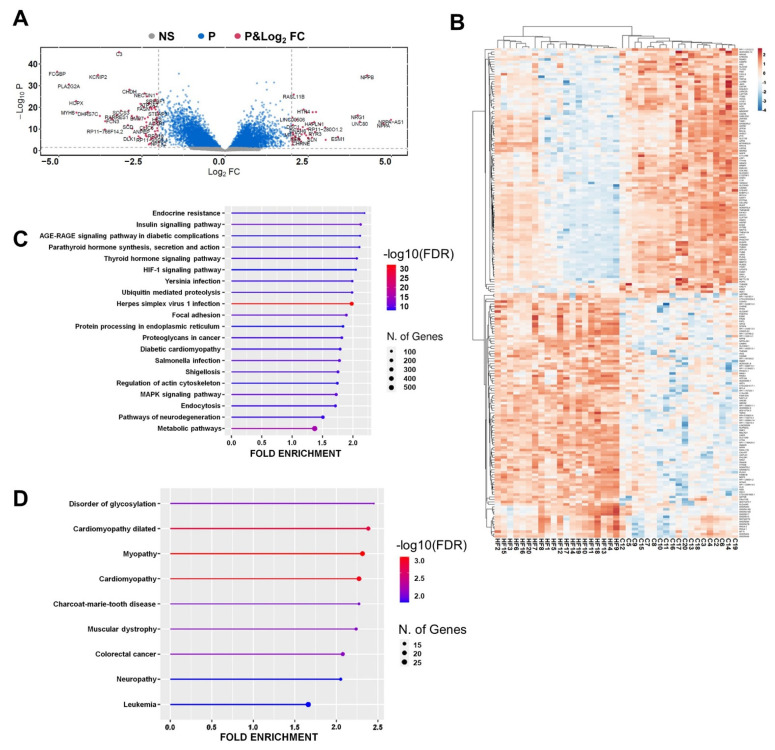
Figure 2.
mRNA is differentially expressed in non-end-stage IHF patients. (A) Volcano plot of DE mRNAs between IHF and controls Vertical and horizontal dotted lines delimit red plots (p and log2 fold change) representing DE mRNAs with padj < 0.01 and log2 Fold Change >|1.0|, blue (P) plots representing mRNAs displaying a significant DE with padj ≥ 0.01 but with a log2 fold change < |1.0, and grey (NS) displaying no significant DE. (B) Heatmap showing each patient and control values expressed as Pearson’s correlation coefficient. The unsupervised clustering showed a perfect segregation of IHF from control samples. (C,D) Enriched top 20 KEGG pathways (C) and enriched OMIM diseases (D) from IHF DE mRNAs are represented by lollipop graphs. The size of the lollipop indicates the number of genes, and colors indicate the statistical significance (red for lower FDR, blue for higher FDR). For details on DE mRNAs, please refer to Supplementary Table S4A.