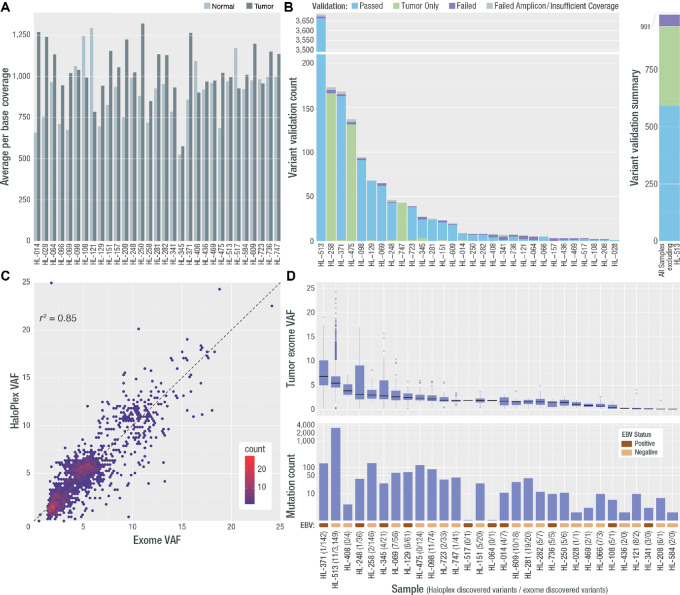
FIGURE 1.
Deep exome coverage, validation rate, final variant count, and VAF per sample. A, Average per base coverage for targeted regions across each tumor and normal sample. B, A targeted orthogonal sequencing strategy (HaloPlex; Agilent) was used to validate all variants that passed filtering strategies for all samples (except for the hypermutated sample, for which a subset of variants were selected for validation—see Materials and Methods). The count of assayed variants that passed, failed, or were validated only in the HaloPlex tumor sample, is shown by sample as well as the number for which validation was not possible due to low tumor coverage or amplicon design failure. The overall count of variants that passed, failed, or received a “tumor only” validation status, excluding variants from HL-513 is shown at the right. Note: One patient (HL-584) did not have any variants pass filtering and review, therefore 30/31 patients were included in validation. C, Comparison of exome VAF and HaloPlex VAF is shown for samples with two or more variants. D, VAF and variant count for all variants across all samples used in all further analyses. Following each sample number is the number of HaloPlex discovered variants and exome discovered variants. In addition, a colored rectangle indicating the EBV stats (assessed using competitive alignment) is shown. Note: Variants from one patient failed validation (HL-157) and no further variants were called in de novo exercises. This patient was removed from all further analyses. In addition, the patient who did not have variants to validate, gained two variants in the de novo exercise and was included in the final cohort (HL-584).