Fig 1.
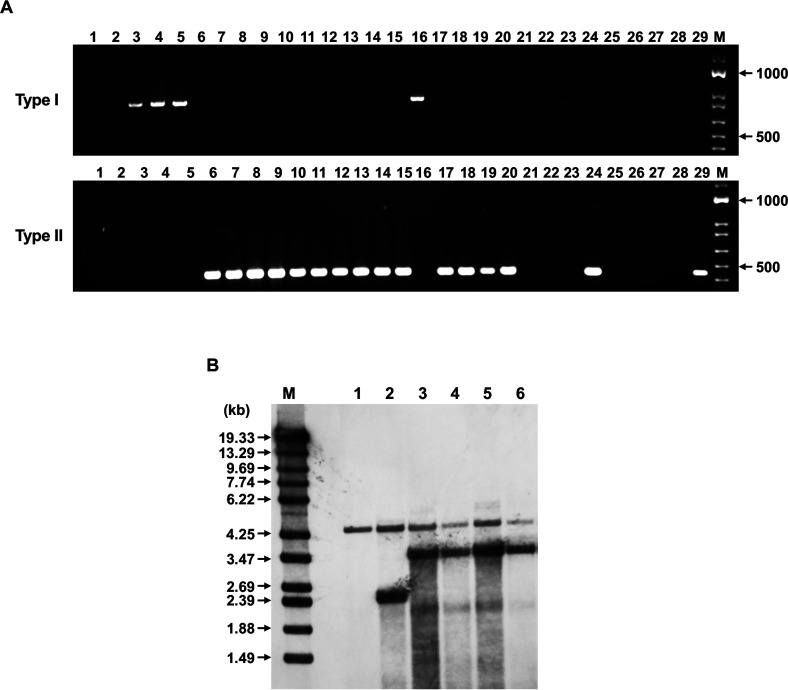
Identification of types I and II strains in T. indotineae. (A) Types I and II strains were identified by amplification of genomic DNA with primer pairs P1–P2 and P3–P4, respectively. TIMM200114 and TIMM 200115, which are susceptible, were used as negative controls (lanes 1 and 2). Azole-resistant strains TIMM200116, TIMM200118, and TIMM 200119, which are known to harbor 5–7 alleles of CYP51B in as many 2,404 bp blocks in tandem, were used as positive controls for the type I strains (lanes 3–5). Lanes 6–29: Search for types I and II tandem sequences in the T. indotineae strains in Table S1. Lanes 6–20: Strains deemed to be resistant to azoles (ITC MIC ≥ 0.25 µg/mL; VRC MIC ≥ 0.25 µg/mL) marked by an asterisk (*) in Table S1. Strain 216510/17 was type I (lane 16). All other strains were type II. Lanes 21–29: Strains deemed to be susceptible to azoles (ITC MIC ≤ 0.25 µg/mL; VRC MIC ≤ 0.25 µg/mL) marked with a hash (#) in Table S1. Two of the later strains (UKJ 1673/17 and 250063/18) were of type II (lanes 24 and 29). (B) Southern blotting analysis of genomic DNA samples from six T. indotineae strains (TIMM20114, TIMM20118, TIMM20120, TIMM20121, TIMM20122, and TIMM20123). Genomic DNA from each strain was digested with XhoI and separated by electrophoresis on a 0.8% (w/v) agarose gel. Lanes 1–6: genomic DNA samples from TIMM20114, TIMM20118, TIMM20120, TIMM20121, TIMM20122, and TIMM20123, respectively. An internal fragment (about 410 bp) of the TinCYP51B gene was amplified by PCR with P21-P16 primers (Table S2) and used as a hybridization probe. The DNA standard fragment sizes are shown on the left.