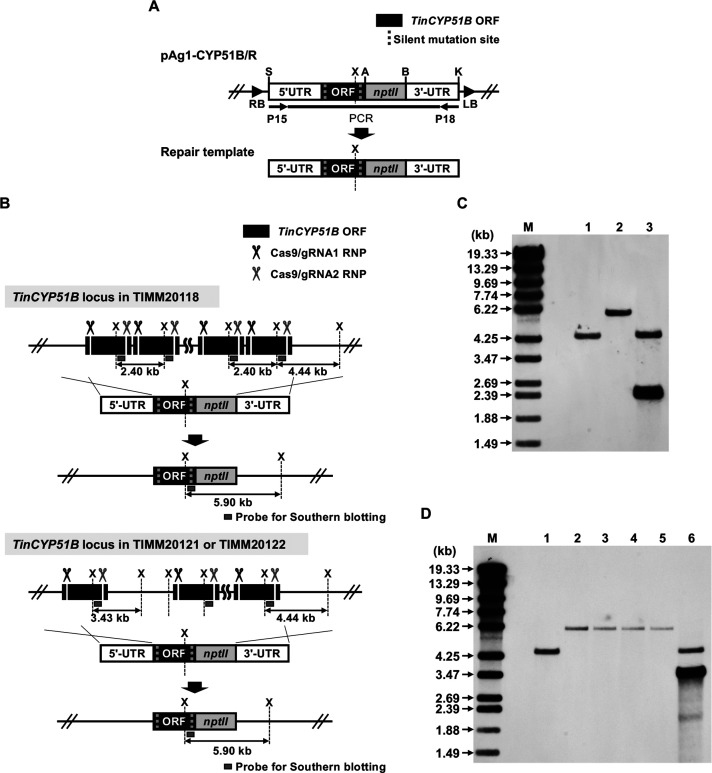
Fig 4.
CRISPR/Cas9-mediated genetic modification of the TinCYP51B locus in the type I azole-resistant strain TIMM20118 and the type II azole-resistant strains TIMM20121 and TIMM20122. (A) Schematic representation of the binary vector pAg1-TinCYP51B/R. The nptII cassette is composed of the promoter sequence of Aspergillus nidulans tryptophan C (trpC) gene (PtrpC), nptII, and the termination sequence of A. fumigatus cgrA gene (TcgrA). The two silent mutations on the repair template cannot be cleaved by the RNP nuclease. Plasmid DNA of pAg1-TinCYP51B/R was used as a template for PCR with P15–P18 primers, to amplify the sequence (repair template) indicated by the thick line. (B) Schematic representation of the TinCYP51B locus before and after modification. The cleaved TinCYP51B locus is replaced by the repair template, resulting in the introduction of the desired modification. The replaced genomic DNA cannot be cleaved by the RNP nuclease. (C AND D) Southern blotting analysis. Aliquots of approximately 5 µg of genomic DNA from each strain were digested with XhoI and separated by electrophoresis on 0.8% (w/v) agarose gels. (C) Lane 1, TIMM20114 (azole susceptible); lane 2, 18 MM_25; lane 3, TIMM20118 (parent strain); M, DNA standard fragments (λ-EcoT14I/BglII digest). (D) Lane 1, TIMM20114 (azole susceptible); lanes 2 to 5, 21 MM_16–1, 21 MM_16–2, 21 MM_23–12, and 21 MM_23–13; lane 6, TIMM20121 (parent strain); M, DNA standard fragments. DNA standard fragment sizes are shown on the left. An internal fragment (about 410 bp) of the TinCYP51B gene was amplified by PCR with P21–P16 primers (Table S2) and used as a hybridization probe.