ABSTRACT
In a looming post-antibiotic era, antibiotic alternatives have become key players in the combat against pathogens. Although recent advances in genomic research allow scientists to fully explore an organism’s genome in the search for novel antibacterial molecules, laborious work is still needed in order to dissect each individual gene product for its antibacterial activity. Here, we exploited phage-induced bacterial morphological changes as anchors to explore and discover a potential phage-derived antimicrobial embedded in the phage genome. We found that, upon vibriophage KVP40 infection, Vibrio parahaemolyticus exhibited morphological changes similar to those observed when treated with mecillinam, a cell wall synthesis inhibitor, suggesting the mechanism of pre-killing that KVP40 exerts inside the bacterial cell upon sieging the host. Genome analysis revealed that, of all the annotated gene products in the KVP40 genome that are involved in cell wall degradation, lytic transglycosylase (LT) is of particular interest for subsequent functional studies. A single-cell morphological analysis revealed that heterologous expression of wild-type KVP40-LT induced similar bacterial morphological changes to those treated with the whole phage or mecillinam, prior to cell burst. On the contrary, neither the morphology nor the viability of the bacteria expressing signal-peptide truncated- or catalytic mutant E80A- KVP40-LT was affected, suggesting the necessity of these domains for the antibacterial activities. Altogether, this research paves the way for the future development of the discovery of phage-derived antimicrobials that is guided through phage-induced morphological changes.
KEYWORDS: antimicrobial screening, bacterial cytological profiling, mechanism of pre-killing, bacteriophage, antimicrobials, lytic transglycosylase
INTRODUCTION
Bacterial infection is among the most concerning problems worldwide. Since the first discovery of antibiotics in the 20th century, antibiotics have represented the best therapeutic solution in the treatment of bacterial infections; however, due to the misuse of antibiotics, many bacteria have somehow genetically adapted to survive through antibiotic treatments (1). The rise of multidrug-resistant (MDR) bacteria has, therefore, become a critical medical issue in human wellness. It has been estimated that from the year 2050 onward, more than 10 million people will die from these MDR pathogens (2). The antibiotic resistance genes in MDR bacteria are even more worrisome since they would lead to unsuccessful treatments for bacterial infections in humans as the antibiotic resistance gene can be transferred (3). If this trend of antimicrobial resistance continues to proceed, the outbreaks associated with the MDR pathogens would soon become too complicated to handle.
Bacteriophage or phage, a prokaryotic virus that replicates inside and kills the specific bacterial host cells, has recently been revisited for the potential as an alternative therapeutic solution for bacterial treatment (4). Phage therapy has been proven successful in many clinical cases in which patients were infected with MDR bacteria, including Pseudomonas aeruginosa, Mycobacterium abscessus, and Acinetobacter baumannii (5 – 7). However, due to the narrow spectrum of host range, inappropriate lysogenic life cycle, and undesired and unidentified genes in the phage genomes, therapy using whole phage particles has extensively raised concerns (8). In addition, the emergence of phage-resistant bacteria might occur when a single-phage treatment is used (9), rendering the phage therapy soon ineffective. Throughout the reproduction cycle, phages produce various phage-derived components for their own benefit that are utilized in hijacking the host fundamental cellular machinery necessary for cell growth and in destabilizing the bacterial cell wall and cell membrane for phage egress. Therefore, these phage-derived proteins have been in the spotlight recently as they are also considered another potential tool through phage therapy. For example, phage-derived enzymes such as virion-associated peptidoglycan hydrolases, polysaccharide depolymerases, and endolysins are becoming the new key in therapy as they serve a crucial role in penetrating the cell wall during phage adsorption and phage-releasing steps (10, 11), thus spearheading the bactericidal capacity in lysing bacterial cells. Since the activity of these enzymes for therapeutic use is limited in Gram-negative bacteria because the enzyme target (peptidoglycan) is hindered by the outer membranes (12 – 14), in recent years, various endolysins were modified to fuse with the bacterial cell wall-binding tags or LPS-destabilizing peptides in order to improve the efficacy against and broaden the specificity for Gram-negative bacteria (13). Due to the fact that there has been an increasing number of unidentified proteins explored within the phage genomes, these hypothetical proteins would be an untapped resource for novel antimicrobial discovery.
With the advancement of omics-related tools nowadays, various phage-derived antimicrobials have been discovered. In 2004, through the bacteriophage genomics of 26 Staphylococcus aureus phages and a phage-mediated bacterial growth inhibition assay, Liu et al. identified 31 novel antimicrobial families, among which ORF104, encoded by phage 77, exerts strong bacteriostatic activity by targeting bacterial DNA replication machinery through DnaI (15). Through interactomics and mass spectrometry, Bossche et al. successfully identified eight phage-derived antimicrobials from lytic Pseudomonas aeruginosa phages, some of which have been confirmed to interfere with the RNA transcription process through interaction with the alpha subunit of the RNA polymerase (16). Moreover, through functional investigation of phage ORFans that are early-expressed small (less than 750 nucleotides), unknown genes in many P. aeruginosa phages by the same research group, they have found various phage-derived proteins exhibiting antimicrobial activities against the bacteria and targeting different global metabolic pathways of the bacteria (17 – 19). During the expression of individual proteins in bacterial cells, some of these proteins trigger substantial morphological changes in bacteria into filamentous cells, such as gp18 from phage LUZ7 (1717) and gp9 derived from phage LUZ24, which has later been identified as a gyrase inhibitor (19).
In addition to genomics, proteomics, and interactomics approaches, phenotypic screening for phage-derived antibacterial molecules is also considered one of the promising strategies. It is well documented that, prior to bacterial cell burst, phage could also induce bacterial morphological changes during infection cycles (17, 18, 20 – 23). For example, the morphological change of bacterial cells from a rod-shaped to a round-shaped cell has been observed during infection with Escherichia coli phage T7. Moreover, PhiKZ-like viruses, also known as the nucleus-forming jumbophages, which are believed to display a complicated infection machinery that is spatiotemporally regulated throughout infection, induce multiple morphological changes through time (23 – 27). Even though host nucleoid morphology cannot be observed since the host chromosome is continuously digested during the infection, the phage triggers cell bulging at the cell center where the phage compartment is located through an unknown mechanism, possibly interfering with cell division components. We recently applied a phenotypic-based antibiotic mechanism of action study, bacterial cytological profiling (BCP), to reveal which cellular pathway of the bacteria is inhibited during the phage infection, the mechanism of pre-killing (MOK) (22). Thammatinna et al. showed that a novel vibriophage named Seahorse induced the formation of host nucleoids toward the signature toroidal-shaped DNA, which is similar to the phenotypic change when cells are treated with tetracycline, a protein translation inhibitor, suggesting the presence of phage-derived proteins that interfere with host protein synthesis during infection.
Even though these signature morphological changes could serve as a starting point in the search for antibacterial molecules embedded in the phage genome, information regarding the direct connection between morphological changes induced by phage and the responsible phage-derived antibacterial molecule is still lacking. Here, we demonstrate how MOK of phage can be used as a guide toward identification and characterization of potential phage-derived antibacterial molecules. First, a single-cell level infection assay revealed that the morphological changes of Vibrio parahaemolyticus EB101 upon the giant vibriophage KVP40 infection are similar to those treated with a cell wall synthesis inhibitor, suggesting that the MOK of KVP40 interferes with host cell wall synthesis. Next, through KVP40 genome analysis, we then selected possible phage-derived candidates that were annotated to be responsible for the observed cell wall synthesis inhibition phenotype. Of all candidates, we found that lytic transglycosylase (LT) was able to induce such a morphological change, suggesting a possible connection between LT and the phage MOK in cell wall synthesis inhibition. Then, genetic-, cytological-, and biochemical-based assays were applied to confirm the mechanism of action of LT as a cell wall synthesis inhibitor and its bactericidal activity. Lastly, we proposed a model for MOK-guided antimicrobial screening in which the pipeline would expedite the process of discovering novel antimicrobial molecules derived from bacteriophages.
RESULTS
KVP40-infected V. parahaemolyticus cells became round prior to cell burst
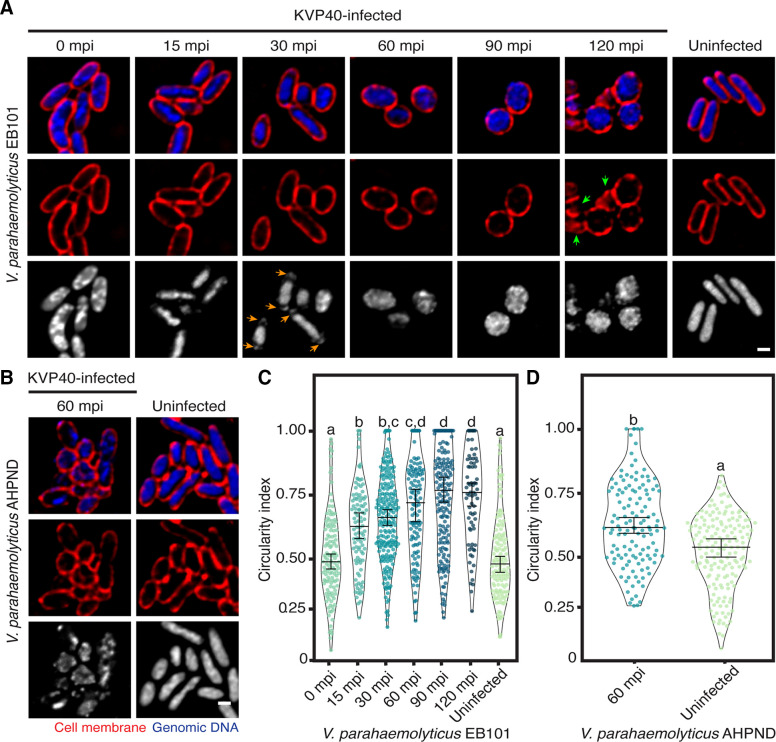
Even though it has been long known that phages randomly replicate inside the host cells, recent findings suggest that the replication machinery of some phages is spatio-temporally regulated at the subcellular level throughout the infection (24, 28). Among them, the PhiKZ-like viruses, also known as nucleus-forming jumbophages, display a complicated infection machinery as they assemble a compartment that encloses phage DNA and partition proteins according to function (26, 27). Due to its large genome (more than 200 kbp), phage KVP40 is also classified as a giant bacteriophage and is assumed to also display organized replication processes (29). However, its replication machinery inside the bacterial cell has never been examined. To explore the infection mechanism of phage KVP40, we performed a single-cell infection assay to observe the morphological changes of V. parahaemolyticus strain EB101 as triggered by the phage KVP40 infection. We observed that, at 0 min post-infection (mpi), the cells were slightly enlarged, followed by the shrinkage of the rod-shaped vibrio cells from 15 to 60 mpi (Fig. 1A; Fig.S1 ). The nucleoid became more condensed as the infection progressed (Fig. 1A; 0–30 mpi). The presence of DAPI-staining puncta at cell poles was visualized at 30 mpi (Fig. 1A; orange arrows), suggesting DNA encapsidation of phage KVP40 during phage maturation (30). Obviously, the infection continuously triggered the bacterial morphological change from a rod shape with a circularity index of 0.5 ± 0.17 at 0 mpi (n = 126) toward a rounded shape with a circularity index of 0.65 ± 0.17 at 30 mpi (n = 215) and 0.74 ± 0.18 at 120 mpi (n = 73) (Fig. 1A; 0–120 mpi, and 1c). We observed no evidence of perfectly rounded cells in the uninfected, control cells (Fig. 1A through D). The infected cells at late time points were visualized as a rounded structure in a cross-section image and appeared as a spherical shape in the Z-stacks (Fig. 1A; 90 and 120 mpi). While a majority of cells remained rounded at 120 mpi, cell lysis was observed, indicating that the reproduction process of the phage has been completed (Fig. 1A; 120 mpi, green arrows). This morphological change of phage-infected cells to a rounded shape was also perceived in other V. parahaemolyticus strains at 60 mpi (Fig. 1B and D; Fig. S1). Altogether, these findings suggested that, through a conserved, unknown infection mechanism, phage KVP40 is capable of inducing phenotypic changes in the infected cells across different strains of the genus Vibrio.
Fig 1.
Unknown mechanism of pre-killing of phage KVP40-induced morphological change of infected cells toward a rounded shape at late infection. V. parahaemolyticus strain EB101 (A) and AHPND (B) were grown in liquid media to an early log phase and were infected by phage KVP40 at MOI 5. At desired time points, the cells were collected and fixed. Prior to observing by microscopy, the cell membrane and nucleoid were stained with FM4-64 (red) and DAPI (blue or gray), respectively. (A) Time-series images over a course of infection (0–120 mpi) of V. parahaemolyticus strain EB101 by phage KVP40. The nucleoid is condensed during 15–30 mpi, followed by the presence of bright DNA-staining puncta close to the cell poles (orange arrows). The cells become rounded after 60 mpi and lyse at 120 mpi (green arrows). (B) Still images of V. parahaemolyticus strain AHPND infected with phage KVP40 at 60 mpi. Scale bars in (A) and (B) equal 1 µm. Graph showing circularity index of (C) V. parahaemolyticus strain EB101 and (D) AHPND cells infected with phage KVP40. Data were collected at the indicated time points from at least three different fields and represented as means ± SEM (n of EB101; 0 mpi = 126, 15 mpi = 100, 30 mpi = 215, 60 mpi = 120, 90 mpi = 172, 120 mpi = 73, and uninfected = 133, and n of AHPND; 60 mpi = 130 and uninfected = 166). Statistical analysis was determined by one-way ANOVA followed by Tukey’s post-hoc tests (P < 0.05).
Mecillinam-treated cells become rounded similar to those infected with KVP40
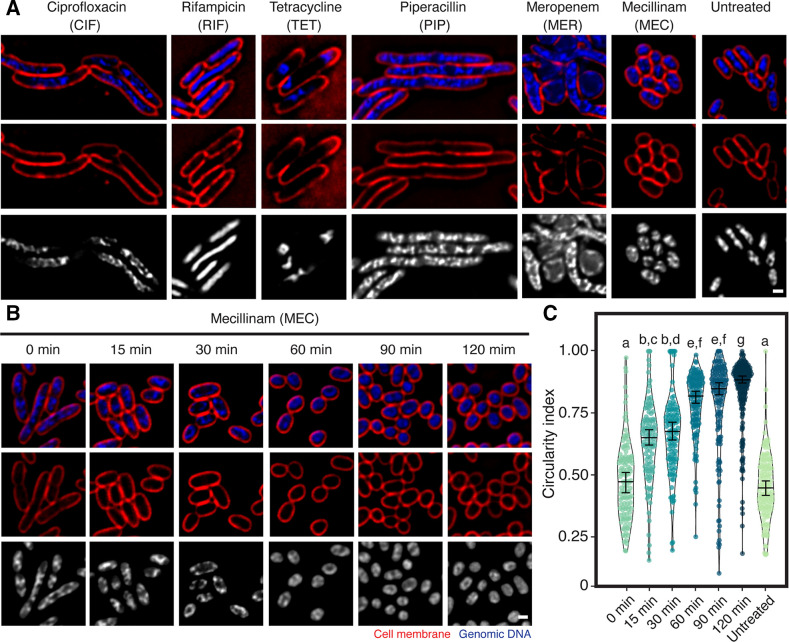
The finding of KVP40-induced morphological changes of the bacteria cells urged us to further determine which cellular pathway of the bacteria was interfered with during infection by comparing with the morphological changes caused by antibiotic perturbation as described in the previous study of mechanism of pre-killing (MOK) (22, 23). To investigate a possible MOK that phage KVP40 exhibits during infection, we selected antibiotics that affect major cellular pathways, including ciprofloxacin for DNA replication, rifampicin for RNA transcription, tetracycline for protein translation, and three different drugs (piperacillin, meropenem, and mecillinam) that target bacterial cell wall synthesis. Upon treatment with the antibiotics, the cell morphology of V. parahaemolyticus was substantially altered (Fig. 2A). Ciprofloxacin-treated cells showed cell elongation and clustered nucleoid at the midcell, while rifampicin and tetracycline caused decondensation and condensation of the bacterial nucleoid, respectively (Fig. 2A; CIF, RIF, and TET). These morphological changes induced by antibiotics in V. parahaemolyticus were consistent with the signature phenotypes previously described in other bacterial species (31, 32), indicating that this morphological-based assay was also applicable to Vibrio spp. Interestingly, concerning the treatment with cell wall inhibitors, almost all vibrio cells treated with mecillinam became rounded (Fig. 2A; MEC, 2b, and 2c) while, upon the meropenem treatment, although the majority of cells were also rounded, some rod-shaped cells remained (Fig. 2A; MER). However, piperacillin, which inhibits different molecular targets from mecillinam and meropenem, induced a different morphological change, resulting in elongated cells with nucleoids distributed along the cells (Fig. 2A; PIP) (33).
Fig 2.
Bacterial cytological profiling of V. parahaemolyticus. Bacteria cells treated with antibiotics targeting different major cellular pathways show different morphological changes (A). Bacterial cells grown until the log phase were treated with 5x MIC of each antibiotic (piperacillin, meropenem, mecillinam, ciprofloxacin, tetracycline, and rifampicin) for 1 h and then fixed in 4% paraformaldehyde. The fixed cells were stained with fluorescent dyes FM4-64 (red) and DAPI (blue) and subjected to fluorescence microscopy. (B) Time-course images of mecillinam-treated bacterial cells. The scale bar represents 1 µm. (C) Violin plot of circularity index of mecillinam-treated bacterial cells. The number of samples per the condition of V. parahaemolyticus EB101 is 0 min = 119, 15 min = 118, 30 min = 215, 60 min = 116, 90 min = 140, 120 min = 194, and untreated = 126. Data were collected at the indicated time points from at least three different fields and represented as means ± SEM. Statistically significant differences between the infected groups and control, as determined by one-way ANOVA (ANOVA) and following Tukey post-hoc test (P < 0.05).
Since we observe that the phenotypic changes of phage KVP40-infected cells are similar to those of mecillinam-treated cells (Fig. 1A; 60 mpi, and Fig. 2A; MEC), we further investigated whether the bacterial morphology, when treated with the drug, was also gradually altered from the rod-shaped cells to the rounded cells similar to that when infected with the phage. The time-series data revealed that the bacterial cells first appeared as a rod structure and were gradually transformed toward the rounded structure throughout the time, becoming completely circular at 120 min (Fig. 2B). The circularity index was then used as a factor to demonstrate the cell roundness, the number of which increased over time, such as 0.48 ± 0.16 (n = 119), 0.64 ± 0.16 (n = 118), 0.67 ± 0.17 (n = 116), 0.79 ± 0.13 (n = 135), 0.78 ± 0.19 (n = 140), and 0.84 ± 0.14 (n = 194), at 0, 15, 30, 60, 90, and 120 min post treatment, respectively (Fig. 2C). Due to the same temporal phenotypic changes between the phage infection and the drug treatment, these data suggested that, during the phage infection at late time points, phage KVP40 displays its MOK through interfering with the bacterial cell wall synthesis, possibly through a similar mechanism to mecillinam, which subsequently led to the loss of cell integrity and shape alteration.
Lytic transglycosylase is a protein candidate from bioinformatic analysis and functional annotation
Based on the predicted MOK of phage KVP40, which is related to the impairment of bacterial cell wall synthesis during late infection, we set out to explore the phage KVP40 genome for the proteins that serve a role in cell wall degradation. From the double-stranded DNA genome of KVP40 that contains 244,834 bps encoding 381 putative open reading frames (Vibrio phage KVP40; NC_005083), we looked through the genome and manually selected three potential candidates based on the functional annotation. They included a lytic transglycosylase SLT domain protein (LT) KVP40.0279.1 (NP_899526.1), a baseplate hub subunit and tail lysozyme KVP40.0335 (NP_899581.1), and a baseplate wedge subunit containing a lysozyme domain KVP40.0340 (NP_899586.1). However, KVP40.0335 and KVP40.0340 have been proven to be parts of the phage baseplate, serving as puncturing devices for phage entry (34, 35). We, therefore, reasoned that KVP40.0279.1 or KVP40-LT, which is acknowledged as one of the bacteriophage endolysins, might play a crucial role in cell wall degradation during late infection for phage KVP40 egress and potentially trigger the morphological change as observed in the single-cell morphological assay.
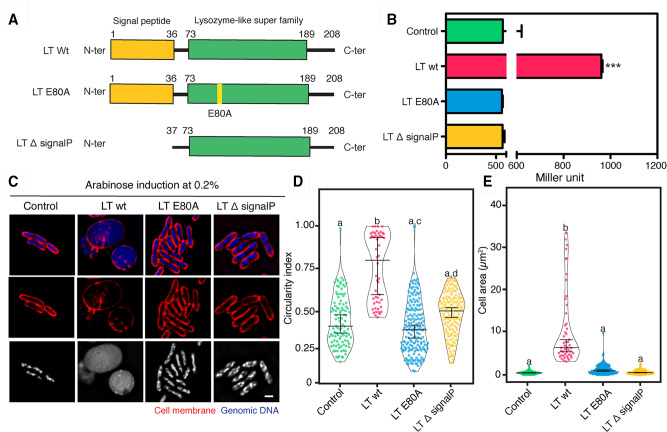
The KVP40-LT gene is 627 bp long and can be translated into a polypeptide with 208 amino acids. The functional amino acid sequence analysis of KVP40-LT by various bioinformatic tools, including BLASTP and INTERPRO, revealed the presence of a conserved protein motif of a lysozyme-like domain (pfam01464) with an E-value score of 1.92e−08 at amino acid positions 73–189, confirming the functional prediction of this protein in the database (Fig. 3A; Fig. S2). Furthermore, the results from the multiple sequence alignment of KVP40-LT with closely related phage LTs demonstrated an extended N-terminal amino acid sequence of the protein that was strongly predicted as a signal peptide at the first 36 amino acids with a score of 0.8637 by SignalP6.0 and a catalytic glutamic residue (E80) in the lysozyme-like domain that is very well conserved across the phage LTs (Fig. 3A; Fig. S2). This E80 of KVP40-LT was well aligned with the catalytic residue E64 that has been previously characterized in chain A of the membrane-bound lytic murein transglycosylase A of E. coli (36), suggesting the critical role of this residue in the transglycosylase activity of KVP40-LT.
Fig 3.
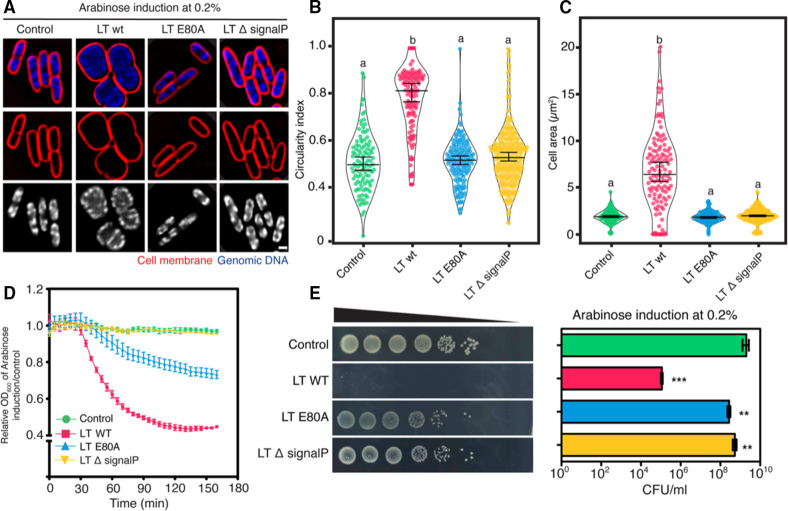
Lytic transglycosylase of phage KVP40 triggers the bacterial morphological change similar to that observed during the phage infection. (A) Illustration of the plasmid constructs used for characterization of the lytic transglycosylase: wild-type LT (LT-wt), the catalytic mutant LT E80A (LT-E80A), and the signal peptide-truncated LT (LT-∆signalP). (B) N-acetylglucosamine assay to determine the activity of lytic transglycosylase in culture of cells expressing LT-wt, LT-E80A, and LT-∆signalP. Data were represented as means ± SD from at least three independent experiments. (C) Single-cell morphological assay showing apparent phenotypes of V. parahaemolyticus cell expressing LT-wt, LT-E80A, and LT-∆signalP. V. parahaemolyticus strain EB101 harboring the indicated construct was grown to an early log phase and the protein expression was induced with 0.2% arabinose. After 10 min of induction, the cells were harvested, fixed, and stained with FM4-64 (cell membrane; red) and DAPI (nucleoid; blue or gray), prior to observing by fluorescence microscopy. Scale bar represents 1 µm. Graphs showing (D) circularity index and (E) cell area of cells expressing LT-wt, LT-E80A, and LT-∆signalP after induction with 0.2% arabinose. Data were collected from at least three different fields and represented as means ± SD (n; control = 102, LT-wt = 50, LT-E80A = 178, LT-∆signalP = 102). Statistical analysis was determined by one-way ANOVA followed by Tukey’s post-hoc tests. The cells harboring the plasmid pBAD33 alone are used as control.
KVP40-LT triggers a morphological change in cells similar to phage KVP40-infected cells
To further confirm the functional annotation of KVP40-LT, we created three constructs, including the wild-type LT, the catalytic-mutant LT E80A, and the signal peptide-truncated LT, and examined the transglycosylase activity of the enzyme and its derivatives in the culture of overexpressing cells by N-acetylglucosamine assay (Fig. 3A and B). The result showed that the transglycosylase activity was detected in the culture of cells expressing wild-type LT at approximately two times greater than those in the culture of cells expressing the catalytic mutant LT E80A and the signal peptide-truncated LT, including the internal control that harbored the vector pBAD33 alone (Fig. 3B). These data indicated that KVP40.0279.1 encodes a fully active KVP40-LT, capable of lysing bacterial cells from inside, and the signal peptide and catalytic residue E80 are necessary for its function.
Since KVP40-LT was assumed to serve a role in phage MOK during late infection and triggers morphological changes to a rounded shape as observed in infected cells, we then performed a single-cell morphological assay by inducing the protein in bacterial cells and visualizing whether the bacterial phenotype was changed in the presence of the protein. As expected, the V. parahaemolyticus cells expressing wild-type LT had their morphology altered to a spherical shape. They were substantially different from the normal rod-shaped cells that expressed the catalytic-mutant LT E80A and the signal peptide-truncated LT (Fig. 3C). Moreover, these cells appeared swollen and were approximately six times larger in size than those that harbored the mutants (Fig. 3D and E). The circularity index as measured in cells expressing wild-type LT was at 0.76 ± 0.2 (n = 50), which was quite similar to the circularity index of infected cells at 120 mpi (Fig. 1C and 3D). Altogether, these findings suggested that KVP40-LT requires the signal peptide and catalytic residue E80 to reach and degrade the bacterial cell wall, subsequently leading to the loss of membrane integrity to maintain turgor pressure in cells and, thus resulting in the bacterial morphological change.
KVP40-LT inhibits V. parahaemolyticus cell growth by lysing the cells
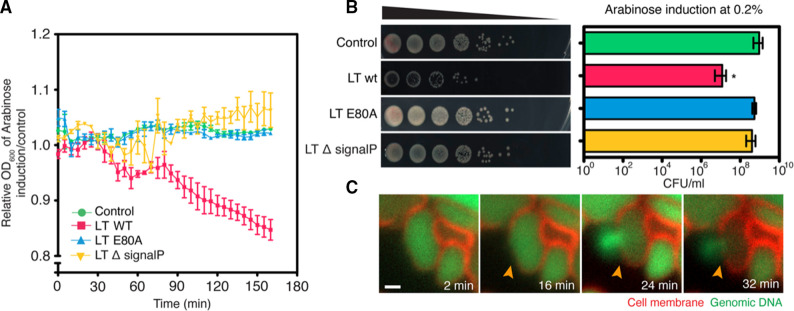
Since we hypothesize that KVP40-LT degrades the bacterial cell wall, leading to the loss of cell integrity by which the turgor pressure can subsequently destroy the cell, we, thus, pursued whether KVP40-LT exhibits bactericidal activity against the bacteria. First, we examined the relative cell density and also analyzed the colony-forming units (CFU) of the V. parahaemolyticus cells expressing the wild-type LT, the catalytic-mutant LT E80A, and the signal peptide-truncated LT, compared to the control cell. Under the induction with 0.2% arabinose, the relative cell density of all groups was unchanged up to 60 min, after which the density of the cells expressing wild-type LT decreased continuously until the 160 min time point (Fig. 4A). However, the cell density in the presence of the mutants remained at the same level as that of the control cells. CFU of cells as measured at 2 h of overexpression revealed that KVP40-LT was able to reduce the bacterial growth around 2-logs while the conditions of removing the signal peptide and mutating the catalytic residue E80 completely impaired the bactericidal activity of the enzyme, further supporting the importance of the signal peptide and catalytic residue E80 in transglycosylase activity (Fig. 4B). The uninduced group showed comparable CFU of bacteria to those of the arabinose-induced groups, indicating no toxicity of arabinose to the cell growth (Fig. S3a). The time-series microscopy data further supported the bactericidal activity of KVP40-LT in a temporal manner. During the expression of KVP40-LT in the cells, V. parahaemolyticus cells initially appeared as a rounded shape, while the bacterial membrane was still intact (Fig. 4C; red). At 16 mpi, the cell membrane was permeabilized, followed by the cell burst at 24 mpi, as indicated by the bright staining of the released genomic DNA (Fig. 4C; green, orange arrows, Video S1). Altogether, these data suggested that KVP40-LT exhibits antibacterial activity through the degradation of the bacterial cell wall, resulting in the loss of cell integrity and eventually leading to cell burst.
Fig 4.
Lytic transglycosylase of phage KVP40 is lethal to V. parahaemolyticus via lysis of the cells from inside out. V. parahaemolyticus EB101 harboring the indicated constructs was grown to an early log phase, and the protein was induced with 0.2% arabinose. The effect of KVP40-LT and its derivative on V. parahaemolyticus was then investigated. (A) Graph showing the OD600 of cells expressing LT-wt, LT-E80A, and LT-∆signalP relative to the control cell. (B) Representative images of growth inhibition assay showing the colony-forming unit (left panel) and a graph (right panel) of cells expressing LT-wt, LT-E80A, and LT-∆signalP. Data in the graph were represented as means ± SD from at least three independent experiments. Statistical analysis was determined by one-way ANOVA followed by Tukey’s post-hoc tests. (C) Time-lapse imaging of V. parahaemolyticus EB101 expressing LT-wt over an interval of 32 min. The cells were grown to an early log phase and inoculated onto agarose pads, supplemented with 0.2% arabinose, FM4-64 (membrane; red), and SYTO 16 (DNA; green). Then, the cells were visualized by fluorescence microscopy. The cells harboring the plasmid pBAD33 alone are used as control. Scale bar represents 1 µm.
KVP40-LT bactericidal mechanism is conserved across bacterial genus
The bacterial cell wall is a rigid structure that maintains the cell shape and structural integrity. It is composed of a complex mesh-like structure called peptidoglycan and is highly conserved among bacteria. As KVP40-LT targets this conserved structure in bacteria, we further investigated if KVP40-LT can also trigger morphological changes in other bacteria and exhibit antimicrobial activity against it. We, therefore, again performed a single-cell morphological assay by inducing KVP40-LT and its derivatives in cells of E. coli, Salmonella enterica Typhimurium, and Pseudomonas aeruginosa and visualized whether cell morphology was altered in the presence of the protein. Similar to V. parahaemolyticus, wild-type LT was able to trigger the phenotypic change of E. coli cells toward a rounded shape with the circularity index at 0.78 ± 0.17 (n = 123) and the cell size being three times larger than the other groups (Fig. 5A through C). The cell shape and size remained unchanged in the cells expressing mutants and appeared in the normal rod-shaped structure similar to the control E. coli cells (Fig. 5A through C). Similarly, wild-type LT also induced the rod-shaped cells of S. enterica Typhimurium and P. aeruginosa toward rounded structures (Fig. S4a and b). However, unlike V. parahaemolyticus, the DAPI-staining nucleoids in E. coli were not diffused throughout the cells but localized as bright puncta close to the cell membrane. This result demonstrated that, due to the similar morphological changes among the tested bacteria in the presence of KVP40-LT, the bactericidal mechanism of how KVP40-LT interfered with the molecular target in bacteria is likely the same.
Fig 5.
Conserved mechanism in bacterial cell lysis of KVP40-derived lytic transglycosylase. E. coli JP313 harboring the indicated constructs was grown to an early log phase prior to protein induction with 0.2% arabinose. The effect of KVP40-LT and its derivative on E. coli was then investigated. (A) Single-cell morphological assay showing apparent phenotypes of E. coli cell expressing LT-wt, LT-E80A, and LT-∆signalP. E. coli harboring the indicated construct was grown to an early log phase and the protein expression was induced with 0.2% arabinose. After 10 min of induction, the cells were harvested and fixed. Cell membrane and nucleoid were stained with FM4-64 (red) and DAPI (blue or gray), prior to observing by microscopy. Scale bar represents 1 µm. Graphs showing (B) circularity index and (C) cell area of E. coli cells expressing LT-wt, LT-E80A, and LT-∆signalP after induction with 0.2% arabinose. Data were collected from at least three different fields and represented as means ± SD (n; control = 108, LT-wt = 123, LT-E80A = 146, LT-∆signalP = 276). Statistical analysis was determined by one-way ANOVA followed by Tukey’s post-hoc tests. (D) Graph showing the OD600 of E. coli cells expressing LT-wt, LT-E80A, and LT-∆signalP relative to the control cell. (E) Representative images of growth inhibition assay showing the colony-forming unit (CFU) (left panel) and a bar graph (right panel) of E. coli cells expressing LT-wt, LT-E80A, and LT-∆signalP. Data in the graph were represented as means ± SD from at least three independent experiments. Statistical analysis was determined by one-way ANOVA followed by Tukey’s post-hoc tests.
To further explore if KVP40-LT also exhibits antimicrobial activity against E. coli, we investigated the reduction of cell density and CFU of the E. coli cells expressing the wild-type LT and the mutants. The result showed that the relative cell density to the control cell during expression of wild-type LT decreased sharply after 30 min of protein induction similar to what was observed in V. parahaemolyticus (Fig. 5D). Even though the density of cells expressing the signal peptide-truncated LT was comparable to that of the control cells, we also observed a declining trend in the density of cells expressing the catalytic mutant LT E80A (Fig. 5D). The growth inhibition assay further confirmed the bactericidal activity of KVP40-LT as it reduced the CFU of E. coli up to 4 logs compared to the cell control. We also observed a slight decrease in CFU of cells expressing the catalytic-mutant LT E80A and the signal peptide-truncated LT (Fig. 5E). The CFU of the uninduced group was comparable to those of the arabinose-induced groups, indicating no toxicity of arabinose to the growth of E. coli (Fig. S3b). As compared to the growth inhibition of KVP40-LT in V. parahaemolyticus, the enzyme displayed more effectiveness in bactericidal activity against E. coli, as it lysed the cells in a shorter time and reduced a higher number of viable cells. This finding suggested that even though the peptidoglycan cell wall is conserved, the membrane components of these bacteria that might make the cells tolerate turgor pressure might be diverse.
DISCUSSION
Morphology-guided antimicrobial discovery has been widely used and successfully determined the mechanism of action (MOA) of the compound of interest against the bacteria (37, 38). Through the morphological changes of bacterial cells upon treatment with various antibiotics, BCP correlates these changes with the MOA of the antibiotics and is, thus, able to identify the MOAs of the compounds (31, 32, 38). Similarly, phages are known to trigger phenotypic alterations in the bacterial host while replicating inside the host cells. During infection, phage encodes a variety of proteins, some of which interact molecularly with targets that are necessary for bacterial growth, contributing to the alteration of cell shape or bacterial nucleoid (22). Thus, based on this similarity, we recently applied the BCP principle to reveal the mechanism of pre-killing (MOK) of a vibriophage in which it exerts to hijack or interfere with the bacterial protein synthesis before phage egress (22). In this study, however, vibrophage KVP40 infection showed none of the previously reported signature phenotypic changes during the early timepoint of infection when bacterial cellular pathways were hijacked by the phage. It is possible that, due to the large genome of phage KVP40 that encodes many phage proteins in the host cell (29), the apparent morphology would be a mixed phenotype that results in multiple MOKs that simultaneously interfere with the host. For example, the nucleus-forming jumbophages that also possess genome sizes larger than 200 kb trigger massive morphological changes in the bacterial host during infection, including cell bulging at midcell where the phage nucleus is located through, an unknown mechanism (30). Therefore, this could make it hard to pinpoint what specific pathway is targeted by the phage.
However, during late infection, the phenotype of infected cells turned toward a rounded structure, this alteration being highly conserved as it also occurs in other strains of V. parahaemolyticus during the phage infection. The shape of the phage KVP40-infected and LT-expressing cells was found to be similar to that of the mecillinam-treated cells, indicating that the same cellular pathway of the bacteria was affected. In this study, cells treated with PIP, MER, and MEC resulted in elongated, bulging, and spherical cells, respectively. These differences in morphological effect are due to the varying affinities toward different penicillin-binding proteins (PBPs) of each antibiotic. For example, MEC is known to directly inhibit the PBP2 of the bacteria (39) which is responsible for lateral cell wall synthesis (40) and elongation (41); inhibiting the enzyme will result in round-shaped cell. We speculate that the spherical shape caused by KVP40 infection and LT expression is not the direct result of PBP2 inhibition but rather the impaired cell wall structure resulting from LT activity at the late stage of infection (42). Since the cell can no longer maintain its regular structure due to the extensive cell wall degradation, turgor pressure exerted by the osmotic flow will contribute to the change in structural integrity (43). This speculation is partly supported by our observation that the morphological change becomes more obvious only after the appearance of intracellular bright puncta, which is a hallmark of successful DNA encapsidation at the late stage of the lytic cycle (25, 30). In addition, the transition of rod-shaped cells to spherical-shaped cells has also been previously reported to be involved in the host cell lysis in other E. coli during late infection (44). Thus, this evidence indicates that this morphological change might serve as an important role for phage reproduction at the late time point after the phage maturation has been completed, possibly the phage egress and host cell lysis.
During phage egress, it has been well characterized that the phage lysis cassette, composed of holin, endolysin, and spanin, is necessary for host cell lysis by destroying the cell wall and modifying the cell membrane (44, 45). Some processes, such as the pinholin-signal anchor release (SAR) endolysin system, have been reported to trigger bacterial cell morphology toward a spherical shape (44). From our analysis, in addition to the functional annotation of phage KVP40 genome (29) narrowing down toward the proteins in the lysis cassette, we found two genes encoding RZ-like spanin, including KVP40.0086 and KVP40.0087, and a lytic transglycosylase SLT domain protein (KVP40.0279.1), which is one of the endolysins as we named it KVP40-LT. During the expression of wild-type KVP40-LT alone in bacterial cells, it is obvious that it can transform the cell shape toward a spherical shape similar to what we observed during the phage infection. This evidence demonstrates that KVP40-LT serves a major role in degrading the bacterial cell wall and induces phenotypic changes during the phage infection at the late stage of the lytic cycle. However, we were curious if KVP40-LT could function alone without other lysis-related proteins, as we did not identify any genes similar to holin in the genome, even in the genes that are supposed to be related to the lysis cassette. It has been widely accepted that phage lysins are highly divergent, and more than 20% of them are holin-independent lysins that harbor either SAR or a cleavable signal peptide (46). In particular, the signal peptide is recognized by the host Sec secretion system, thus facilitating the transportation across the inner membrane into the cell periplasm (46). Interestingly, according to our analysis, rather than the conserved domain of the lysozyme-like superfamily, KVP40-LT appears to contain a signal peptide that can be recognized by the Sec machinery of bacteria. Therefore, with the presence of a signal peptide on KVP40-LT and the absence of holin in the genome, this finding suggests that KVP40-LT is classified as one of the holin-independent lysins that, by itself, can degrade the bacterial cell wall without the need of holin. Our result clearly supported this assumption since expressing the wild-type KVP40-LT alone in bacterial cells can digest the peptidoglycan and trigger morphological change leading to cell death, while removing the signal peptide and mutating the catalytic residues involved in transglycosylase activity completely restrained the function of the enzyme.
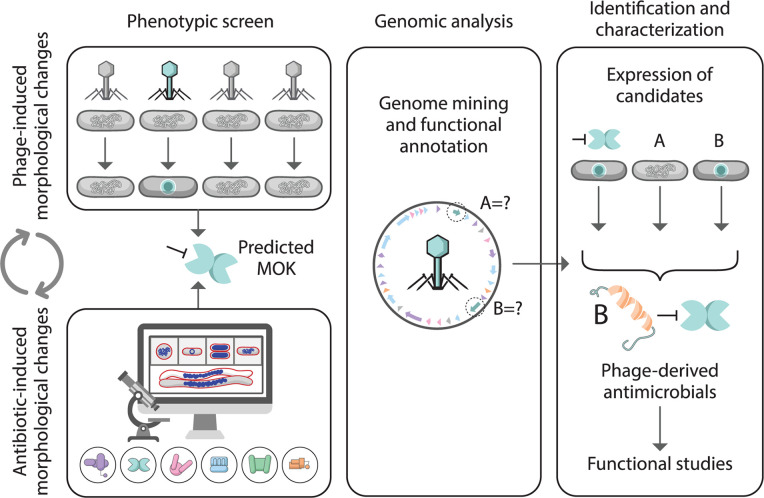
Altogether, our investigation of morphological changes in bacterial cells leads to a phage-derived antimicrobial protein, and thus, we propose our model for phage-induced morphology-guided antimicrobial screening. As illustrated in Fig. 6, we begin by exploring the phages in the collection by a single-cell infection assay and selecting the ones that trigger a morphological change in the bacterial host that matches the signature phenotype on our BCP database. This step will facilitate the narrowing down to only the phages in the library that exhibit interesting MOK for subsequent genome analysis and functional annotation, thus avoiding massively laborious work on individual phage genomic studies. Due to the matched morphology between phage-infected and drug-treated cells, we can then predict the possible MOK that the phage displays during its lytic cycle. Based on the predicted MOK together with the available phage genomics information, we will be guided toward a group of genes in the phage genome that are responsible for this change for further functional investigation, thereby easily ignoring the rest of the genome that are not related to it. However, some limitations of the screening are worth noting here: (i) the method relies mainly on gene annotation; thus, unknown genes that are difficult to bioinformatically annotate might hamper the target selection of the pipeline; (ii) optimization and synchronization of each phage for its infection are needed; (iii) we cannot pinpoint the exact molecular targets which the phage hijacks; and (iv) the morphological changes of bacterial cells during phage infection are quite varied and some do not match the database, which might be the result of the multiple phage MOKs that interfere with bacterial cells simultaneously. Therefore, the fundamentals of how the phage-derived antimicrobials interact with bacterial targets will be needed, and an extension of our BCP database to incorporate a combination of antibiotics will be required to reveal all possible bacterial morphological changes.
Fig 6.
Phage-induced morphology guided antimicrobial screening. Through the matched morphology during phenotypic screening as induced by phages and antibiotics, this pipeline suggests the predicted MOK which the phage potentially displays to interfere with host metabolic processes. Based on the predicted MOK together with genomic analysis of the phage, the information narrows down the selection toward potential candidates that could serve a role in the apparent morphology during the phage infection. In vivo expression of each selected candidate to examine the morphological change is lastly conducted to confirm the role of the protein in phenotypic alteration.
MATERIALS AND METHODS
Bacterial growth and bacteriophage preparation
Vibrio parahaemolyticus strain EB101 and AHPND (Table S1) were used as the host for phage propagation, the single-cell infection assay, and the single-cell morphological assay. Bacterial cells were grown overnight in Tryptic Soy Broth (TSB) (HiMedia Soyabean Casein Digest Medium, Cat. No. M011) supplemented with 1.5% NaCl at 37°C, 250 rpm. E. coli DH5a, E. coli JP313, S. enterica Typhimurium IR715, and P. aeruginosa PA01 (Table S1) were used for molecular cloning and the single-cell morphological assay. The bacterial cells were grown overnight in Luria-Bertani (LB) broth (Tryptone; Himedia, Cat. No. RM027, and Yeast extract; Himedia, Cat. No. RM014) at 37°C, 250 rpm.
Giant vibriophage KVP40 was isolated and identified as previously reported by (47). Briefly, the high-titer phage lysate was prepared using the double-layer agar method. Phage was 10-fold serially diluted in SM buffer and 10 µL of each diluted sample was mixed with 100 µL of an overnight culture of the host (V. parahaemolyticus strain EB101). The mixtures were mixed gently and incubated at room temperature for 5 min to initiate the phage adsorption. After that, 5 mL of 0.35% of top agar was added to each sample, and then, the mixtures were poured onto TSA plate containing 1.5% NaCl. The plates were incubated at 37°C overnight. Five milliliters of SM buffer was then added onto the plate that shows confluent lysis, followed by incubation at room temperature for at least 5 h. The phage solution was collected from the plate and was centrifuged at 9,000 rpm for 5 min to pellet the cell debris. The supernatant was then collected and filtered using a 0.45-µm filter. This solution, called the high-titer phage lysate, was evaluated for the phage titer (pfu/mL) and stored at 4°C until use.
This work has been reviewed and approved by Chulalongkorn University-Institutional Biosafety Committee (CU-IBC) in accordance with the levels of risk in pathogens and animal toxins, listed in the Risk Group of Pathogen and Animal Toxin (2017) published by Department of Medical Sciences (Ministry of Public Health), the Pathogen and Animal Toxin Act (2015), and Biosafety Guidelines for Modern Biotechnology BIOTEC (2016) with approval number: SC CU-IBC-028/2020.
Plasmid constructions and bacterial transformation
The genes KVP40.0279.1 encoding full-length lytic transglycosylase and signal peptide-truncated lytic transglycosylase were amplified from the high-titer phage lysate by polymerase chain reaction using specific primers (Table S2). The gene was cloned into the linearized backbone pBAD33 vector at the downstream region of the arabinose inducible promoter and ribosomal binding site using NEBuilder HiFi DNA assembly Master Mix (Catalog no. E2621S) to generate a recombinant plasmid as listed in Table S1. The recombinant plasmid was later transformed into E. coli DH5a, and the transformants were selected on LB media supplemented with corresponding antibiotics (Table S1). Once the construct of full-length lytic transglycosylase had been obtained, point-mutation at aspartic 80 was performed using specific primers (Table S2) by site-directed mutagenesis PCR protocol to alter aspartic to alanine. The correctness of constructs was confirmed by colony PCR and DNA sequencing. The resulting constructs were later transformed into indicated organisms including E. coli JP313, V. parahaemolyticus EB101, S. enterica Typhimurium IR715, or P. aeruginosa PA01 by electroporation (Table S1). Positive clones were stored in glycerol stock at −80°C until used.
Single-cell infection assay
As previously described by Thammatinna et al. (22), overnight cultures of V. parahaemolyticus strains EB101 or AHPND were 100-fold diluted in fresh LB and incubated at 37°C on a roller until OD600 reached an early log phase (≈0.4). Then, 1 mL of bacterial cells was harvested by centrifugation at 8,000 × g for 5 min and infected with the high-titer bacteriophage at MOI 5. At desired time points: 0-, 15-, 30-, 60-, 90-, and 120 mpi, the infected cells were collected and fixed with paraformaldehyde at a final concentration of 4%. Then, the fixed cells were washed three times in 1× PBS and stained with fluorescent dyes; FM 4–64 (1 μg/mL) and DAPI (2 µg/mL) for 10 min. The stained cells were later loaded onto an agarose pad (1.2% agarose with 20% LB) on concave glass slides, and fluorescence microscopy was then performed to observe the morphologies of the infected cells.
Mechanism of action study based on BCP principle
As described by Htoo et al. (31), overnight cultures of V. parahaemolyticus were diluted 1:100 and grown at 37°C until the early log phase. Antibiotics (piperacillin, meropenem, mecillinam, ciprofloxacin, tetracycline, and rifampicin) were added at 5× MIC, incubated for 1 h, and then fixed in 4% final concentration of paraformaldehyde. Fixed cells of treated V. parahaemolyticus were stained with 1 µg/mL of FM 4–46 and 2 µg/mL of DAPI for 5 min. Then, the cell pellet was harvested by centrifugation at 6,000 × g for 30 s and resuspended in 1/10 vol of TSB supplement with 1.5% salt. Three microliters of the bacterial culture was loaded onto agarose pads—the preparation of which have been detailed in the previous section—on concave glass slides and then fluorescence microscopy was performed to observe the morphologies of the altered cell changes. For data analysis, the number of cells (n = at least 100 cells) in the infected condition was compared with that of the uninfected control. The data were collected from at least three independent biological experiments. Image analysis and processing were performed on Fiji software (48).
Bioinformatics and functional annotation
Through functional annotation of KVP40 genome as previously reported, potential candidates that appeared to be involved in peptidoglycan degradation were manually selected. For the identification of homologs of lytic transglycosylase (KVP40.0279.1), the amino acid sequence of KVP40 lytic transglycosylase was used as a query against the NCBI database to obtain related viral lytic transglycosylases (49). Multiple amino acid sequence alignments of the obtained sequences were then generated using MEGA with the default settings to demonstrate the conserved domains. The conserved domains of KVP40 lytic transglycosylase were then confirmed by BLASTP and INTERPRO. SignalP was also used to predict the presence of a signal peptide (50).
Lytic transglycosylase activity assay
To functionally prove whether KVP40 lytic transglycosylase exhibits the transglycosylase activity as predicted by bioinformatics, 4-nitrophenyl-n-acetyl-β-d-glucosaminide (NP-GlcNAc) (catalog no. N-230-500), which is a chromogenic substrate, was used as a substrate to evaluate the enzyme activity in the assay. Briefly, V. parahaemolyticus strain EB101 harboring pBAD33 plasmid, LT-WT, LT-E80A, and LT-∆signalP were grown in LB supplemented with chloramphenicol (30 µg/mL) at 37°C overnight. Then, starting from the cultures at the same cell density to normalize the cell number, the cells were induced by 0.2% arabinose and incubated with shaking for 2 h; thereafter, the supernatant of cell cultures was collected by centrifugation at 10,000 × g for 10 min to remove the cell pellet and cell debris. Then, 500 µL of supernatant of each strain was mixed with 100 µL of 5 mM NP-GlcNAc, and the reactions were incubated at 37°C for 30 min. Once completed, the reaction was then terminated by adding 0.5 M Na2CO3 at 1:1 ratio. The absorbance of the reaction was measured at 420 nm to evaluate the degradation of NP-GlcNAc (45).
Single-cell morphological assay
To investigate whether KVP40 lytic transglycosylase affects bacterial morphological change of V. parahaemolyticus strains EB101, E. coli JP313, S. enterica Typhimurium IR715, and P. aeruginosa PA01, overnight cultures of the bacteria harboring the indicated recombinant plasmid (Table S1) were 100-fold diluted in fresh LB media (for E. coli, S. enterica Typhimurium, and P. aeruginosa) and supplemented with 1.5% of NaCl (for Vibrio), and then incubated at 37°C on a roller until OD600 reaches an early log phase. Then, arabinose was added to the culture at 0.2% final concentrations and incubated for 10 min. The bacterial cells were then harvested, fixed, and stained as described above. Three microliters of the sample was loaded onto an agarose pad (1.2% agarose with 20% LB) on concave glass slides followed by fluorescence microscopy.
For live-cell imaging, day cultures of V. parahaemolyticus with OD600 at an early log phase were prepared as described above. The bacterial cells were then transferred onto an agarose pad and induced simultaneously by 0.2% arabinose. The cells were also stained with fluorescent dyes: FM4-64 for membrane staining and the cell-permeant Syto16 (catalog no. S7578) for DNA staining, on concave glass slides. The slides were incubated in a moist chamber with a temperature controller at 37°C, and time-lapse imaging was then initiated with a window of 60 min with 2-min intervals. Images were collected and analyzed by Fiji software.
In vivo killing assay and in vivo growth inhibition assay
To verify the antibacterial effect of KVP40 lytic transglycosylase on bacterial growth, V. parahaemolyticus strains EB101 and E. coli JP313 harboring the indicated recombinant plasmid were grown in 5 mL of liquid LB media supplemented with 30 µg/mL chloramphenicol at 37°C overnight. Overnight cultures were then 100-fold diluted into fresh LB and grown at 37°C until OD600 reached an early log phase (≈0.4). For in vivo killing assay, a final concentration of 0.2% arabinose was added to induce the protein expression in the cultures in experimental groups. The absorbance at OD600 was then monitored for 3 h with 5-min intervals on a microplate-reader while maintaining the temperature at 37°C throughout the experiment. For in vivo growth inhibition assay, the cultures were induced by either 0.2% arabinose (experimental group) or uninduced (control group) for 2 h. Cultures were then 10-fold diluted in fresh LB, and 5 µL of the mixtures was then spotted on LB agar containing 30 µg/mL chloramphenicol. The plates were dried and incubated at 30°C overnight. Colony-forming unit (CFU/mL) was counted on the following day.
Quantitation and data analysis
For data analysis, the number of cells (n = least 100 cells) in the infected condition was compared with that of the uninfected control. The data were collected from at least three independent biological experiments. Image analysis and processing were performed on Fiji software.
For the analysis, raw images from the fluorescence microscope were pre-processed and analyzed on Fiji software. The morphological parameters were selected based on bacterial cell wall. FM4-64 was used to define the outlines for defining the circularity index and area. The background intensity of each cell was subtracted to obtain the boundary of the cell. In order to determine the circularity of bacterial cell wall in infected cells and induced cells, we used the tool "Analyze Particles" in Fiji program as described by Chaikeeratisak et al. (25). The dead cell, cell debris, or unconnected boundary cell was subtracted in this analysis. Statistical analysis was performed, and statistical significance was determined by student’s t-test and one-way ANOVA with Tukey’s HSD post-hoc test. The analyses for each experiment are mentioned in the figure legends. The Violin plot was generated from Plots of Data (https://huygens.science.uva.nl/PlotsOfData/) (51).
ACKNOWLEDGMENTS
This work was financially supported by Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (Grant no. RGNS 63-001), and the Asahi Glass Foundation under CU-AGF grant 2021 (V.C.). P.N. was supported by National Research Council of Thailand (NRCT) and Mahidol University: N42A650368, and the Coordinating Center for Thai Government Science and Technology Scholarship Students (CSTS) – the National Science and Technology Development Agency (NSTDA) grant no. JRA-CO-2563-13729-TH. The student fellowships to T.S. from the 90th Anniversary of Chulalongkorn University Fund through Ratchadaphiseksomphot Endowment Fund: GCUGR1125651038D number 038 are acknowledged. We also acknowledge the additional support from the Ratchadaphiseksomphot Endowment Fund, Chulalongkorn University to the Center of Excellence for Molecular Biology and Genomics of Shrimp and support for scientific instruments by the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA), Science and Technology Research Partnership for Sustainable Development, SATREPS JPMJSA1806 (V.C. and P.N.).
We would like to thank Mahidol University Frontier Research Facility (MU-FRF) for instrumentation support for DeltaVision Ultra.
Contributor Information
Vorrapon Chaikeeratisak, Email: Vorrapon.c@chula.ac.th.
Pranita D. Tamma, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00764-23.
Supplemental figure legends, Tables S1 and S2, and Movie S1 legend
Fig. S1
Fig. S2
Fig. S3
Fig. S4
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Neil J. n.d. Review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. Available from: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf
- 3. Thornber K, Verner-Jeffreys D, Hinchliffe S, Rahman MM, Bass D, Tyler CR. 2020. Evaluating antimicrobial resistance in the global shrimp industry. Rev Aquac 12:966–986. doi: 10.1111/raq.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–232. doi: 10.1016/j.chom.2019.01.014 [DOI] [PubMed] [Google Scholar]
- 5. Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wright A, Hawkins CH, Änggård EE, Harper DR. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol 34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x [DOI] [PubMed] [Google Scholar]
- 7. Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61:e00954-17. doi: 10.1128/AAC.00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance, In Advances in applied Microbiology. Elsevier. doi: 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- 9. Principi N, Silvestri E, Esposito S. 2019. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front Pharmacol 10:513. doi: 10.3389/fphar.2019.00513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernández-Ruiz I, Coutinho FH, Rodriguez-Valera F. 2018. Thousands of novel endolysins discovered in uncultured phage genomes. Front Microbiol 9:1033. doi: 10.3389/fmicb.2018.01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gordillo Altamirano FL, Barr JJ. 2019. Phage therapy in the postantibiotic era. Clin Microbiol Rev 32:e00066-18. doi: 10.1128/CMR.00066-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S, Lee D-W, Jin J-S, Kim J. 2020. Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J Glob Antimicrob Resist 22:32–39. doi: 10.1016/j.jgar.2020.01.005 [DOI] [PubMed] [Google Scholar]
- 13. Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay J-P, Miller S, Volckaert G, Lavigne R. 2014. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5:e01379-14. doi: 10.1128/mBio.01379-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodríguez-Rubio L, Chang W-L, Gutiérrez D, Lavigne R, Martínez B, Rodríguez A, Govers SK, Aertsen A, Hirl C, Biebl M, Briers Y, García P. 2016. 'Artilysation' of endolysin λsa2Lys strongly improves its enzymatic and antibacterial activity against streptococci”. Sci Rep 6:35382. doi: 10.1038/srep35382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. 2004. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol 22:185–191. doi: 10.1038/nbt932 [DOI] [PubMed] [Google Scholar]
- 16. Van den Bossche A, Ceyssens P-J, De Smet J, Hendrix H, Bellon H, Leimer N, Wagemans J, Delattre A-S, Cenens W, Aertsen A, Landuyt B, Minakhin L, Severinov K, Noben J-P, Lavigne R. 2014. Systematic identification of hypothetical bacteriophage proteins targeting key protein complexes of Pseudomonas aeruginosa. J Proteome Res 13:4446–4456. doi: 10.1021/pr500796n [DOI] [PubMed] [Google Scholar]
- 17. Wagemans J, Blasdel BG, Van den Bossche A, Uytterhoeven B, De Smet J, Paeshuyse J, Cenens W, Aertsen A, Uetz P, Delattre A-S, Ceyssens P-J, Lavigne R. 2014. Functional elucidation of antibacterial phage orfans targeting Pseudomonas aeruginosa. Cell Microbiol 16:1822–1835. doi: 10.1111/cmi.12330 [DOI] [PubMed] [Google Scholar]
- 18. Wagemans J, Delattre A-S, Uytterhoeven B, De Smet J, Cenens W, Aertsen A, Ceyssens P-J, Lavigne R. 2015. Antibacterial phage orfans of Pseudomonas aeruginosa phage LUZ24 reveal a novel MvaT inhibiting protein. Front Microbiol 6:1242. doi: 10.3389/fmicb.2015.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Smet J, Wagemans J, Boon M, Ceyssens P-J, Voet M, Noben J-P, Andreeva J, Ghilarov D, Severinov K, Lavigne R. 2021. “The bacteriophage LUZ24 "Igy" peptide inhibits the pseudomonas DNA gyrase”. Cell Rep 36:109567. doi: 10.1016/j.celrep.2021.109567 [DOI] [PubMed] [Google Scholar]
- 20. Yang X, Wisuthiphaet N, Young GM, Nitin N, DebRoy C. 2020. Rapid detection of Escherichia coli using bacteriophage-induced lysis and image analysis. PLoS ONE 15:e0233853. doi: 10.1371/journal.pone.0233853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang P, Zhao X, Wang Y, Du K, Wang Z, Yu J, Chang G, Matthews S, Wang H, Liu B. 2022. Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins. Proc Natl Acad Sci USA 119:e2116278119. doi: 10.1073/pnas.2116278119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thammatinna K, Egan ME, Htoo HH, Khanna K, Sugie J, Nideffer JF, Villa E, Tassanakajon A, Pogliano J, Nonejuie P, Chaikeeratisak V. 2020. A novel vibriophage exhibits inhibitory activity against host protein synthesis machinery. Sci Rep 10:2347. doi: 10.1038/s41598-020-59396-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaikeeratisak V, Nguyen K, Egan ME, Erb ML, Vavilina A, Pogliano J. 2017. The phage nucleus and tubulin spindle are conserved among large pseudomonas phages. Cell Rep 20:1563–1571. doi: 10.1016/j.celrep.2017.07.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaikeeratisak V, Birkholz EA, Prichard AM, Egan ME, Mylvara A, Nonejuie P, Nguyen KT, Sugie J, Meyer JR, Pogliano J. 2021. Viral speciation through subcellular genetic isolation and virogenesis incompatibility. Nat Commun 12:342. doi: 10.1038/s41467-020-20575-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chaikeeratisak V, Nguyen K, Khanna K, Brilot AF, Erb ML, Coker JKC, Vavilina A, Newton GL, Buschauer R, Pogliano K, Villa E, Agard DA, Pogliano J. 2017. Assembly of a nucleus-like structure during viral replication in bacteria. Sci 355:194–197. doi: 10.1126/science.aal2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chaikeeratisak V, Birkholz EA, Pogliano J. 2021. The phage nucleus and PhuZ spindle: defining features of the subcellular organization and speciation of nucleus-forming jumbo phages. Front Microbiol 12:641317. doi: 10.3389/fmicb.2021.641317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaikeeratisak V, Khanna K, Nguyen KT, Egan ME, Enustun E, Armbruster E, Lee J, Pogliano K, Villa E, Pogliano J. 2022. Subcellular organization of viral particles during maturation of nucleus-forming jumbo phage. Sci Adv 8:eabj9670. doi: 10.1126/sciadv.abj9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trinh JT, Shao Q, Guan J, Zeng L. 2020. Emerging heterogeneous compartments by viruses in single bacterial cells. Nat Commun 11:3813. doi: 10.1038/s41467-020-17515-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol 185:5220–5233. doi: 10.1128/JB.185.17.5220-5233.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang H, Nguyen K, Pham DSL, Agard DA, Pogliano J. 2012. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149:1488–1499. doi: 10.1016/j.cell.2012.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Htoo HH, Brumage L, Chaikeeratisak V, Tsunemoto H, Sugie J, Tribuddharat C, Pogliano J, Nonejuie P. 2019. Bacterial cytological profiling as a tool to study mechanisms of action of antibiotics that are active against Acinetobacter baumannii. Antimicrob Agents Chemother 63:e02310-18. doi: 10.1128/AAC.02310-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nonejuie P, Burkart M, Pogliano K, Pogliano J. 2013. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci USA 110:16169–16174. doi: 10.1073/pnas.1311066110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sumita Y, Fukasawa M. 1995. Potent activity of meropenem against Escherichia coli arising from its simultaneous binding to penicillin-binding proteins 2 and 3. J Antimicrob Chemother 36:53–64. doi: 10.1093/jac/36.1.53 [DOI] [PubMed] [Google Scholar]
- 34. Lynch KH, Stothard P, Dennis JJ. 2010. Genomic analysis and relatedness of P2-like phages of the burkholderia cepacia complex. BMC Genomics 11:599. doi: 10.1186/1471-2164-11-599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nemoto M, Mio K, Kanamaru S, Arisaka F. 2008. ORF334 in vibrio phage KVP40 plays the role of gp27 in T4 phage to form a heterohexameric complex. J Bacteriol 190:3606–3612. doi: 10.1128/JB.00095-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Byun B, Mahasenan KV, Dik DA, Marous DR, Speri E, Kumarasiri M, Fisher JF, Hermoso JA, Mobashery S. 2018. Mechanism of the Escherichia coli MltE lytic transglycosylase, the cell-wall-penetrating enzyme for type VI secretion system assembly. Sci Rep 8:4110. doi: 10.1038/s41598-018-22527-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hudson MA, Lockless SW. 2022. Elucidating the mechanisms of action of antimicrobial agents. mBio 13:e0224021. doi: 10.1128/mbio.02240-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Godinez WJ, Chan H, Hossain I, Li C, Ranjitkar S, Rasper D, Simmons RL, Zhang X, Feng BY. 2019. Morphological deconvolution of beta-lactam polyspecificity in E. coli. ACS Chem Biol 14:1217–1226. doi: 10.1021/acschembio.9b00141 [DOI] [PubMed] [Google Scholar]
- 39. Neu HC. 1983. Penicillin-binding proteins and role of amdinocillin in causing bacterial cell death. Am J Med 75:9–20. doi: 10.1016/0002-9343(83)90089-x [DOI] [PubMed] [Google Scholar]
- 40. Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x [DOI] [PubMed] [Google Scholar]
- 41. Pinette MF, Koch AL. 1988. Turgor pressure responses of a gram-negative bacterium to antibiotic treatment, measured by collapse of gas vesicles. J Bacteriol 170:1129–1136. doi: 10.1128/jb.170.3.1129-1136.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scheurwater E, Reid CW, Clarke AJ. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol 40:586–591. doi: 10.1016/j.biocel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 43. Heesterbeek DAC, Muts RM, van Hensbergen VP, de Saint Aulaire P, Wennekes T, Bardoel BW, van Sorge NM, Rooijakkers SHM. 2021. Outer membrane permeabilization by the membrane attack complex sensitizes gram-negative bacteria to antimicrobial proteins in serum and phagocytes. PLoS Pathog 17:e1009227. doi: 10.1371/journal.ppat.1009227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berry J, Rajaure M, Pang T, Young R. 2012. The spanin complex is essential for lambda lysis. J Bacteriol 194:5667–5674. doi: 10.1128/JB.01245-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Zhang C, Wei F, Yu F, Zhao Z. 2021. Bactericidal activity of a holin-endolysin system derived from vibrio alginolyticus phage HH109. Microb Pathog 159:105135. doi: 10.1016/j.micpath.2021.105135 [DOI] [PubMed] [Google Scholar]
- 46. Gontijo MTP, Vidigal PMP, Lopez MES, Brocchi M. 2021. Bacteriophages that infect gram-negative bacteria as source of signal-arrest-release motif Lysins. Res Microbiol 172:103794. doi: 10.1016/j.resmic.2020.103794 [DOI] [PubMed] [Google Scholar]
- 47. Matsuzaki S, Tanaka S, Koga T, Kawata T. 1992. A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol Immunol 36:93–97. doi: 10.1111/j.1348-0421.1992.tb01645.x [DOI] [PubMed] [Google Scholar]
- 48. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dik DA, Marous DR, Fisher JF, Mobashery S. 2017. Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit Rev Biochem Mol Biol 52:503–542. doi: 10.1080/10409238.2017.1337705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, von Heijne G, Nielsen H. 2022. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol 40:1023–1025. doi: 10.1038/s41587-021-01156-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Postma M, Goedhart J. 2019. Plotsofdata-a web app for visualizing data together with their summaries. PLoS Biol 17:e3000202. doi: 10.1371/journal.pbio.3000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure legends, Tables S1 and S2, and Movie S1 legend
Fig. S1
Fig. S2
Fig. S3
Fig. S4