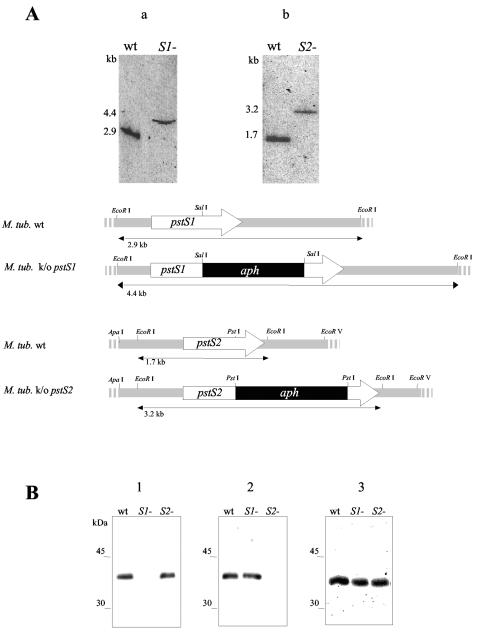
FIG. 1.
(A) Southern blot analysis of the M. tuberculosis (M. tub.) H37Rv pstS1 and pstS2 knockout (k/o) mutants. Genomic DNA was digested with EcoRI, subjected to electrophoresis, blotted onto membranes, and probed with the pstS1 (an NaeI-SalI fragment of the pstS1gene) (a) or the pstS2 (a SacI-PstI fragment of the pstS2 gene) (b) probe. The probes were labeled with [α-32P]dCTP using the Megaprime random-primed labeling kit (Amersham). The hybridization and washing protocols were carried out under high-stringency conditions as described previously (5). The sizes of the hybridizing bands, indicated on the left, were determined from the migration distance of the DNA molecular marker Smartladder (Eurogentec). The arrows depict the lengths and transcriptional orientations of the pstS1 and pstS2 genes. The black boxes represent the aph gene, and the hatched boxes show the pstS1 and pstS2 gene flanking regions used for allelic exchange. Only the relevant restriction sites are indicated. wt, wild type. (B) Immunoblot analysis of the lysates of the pstS1 and pstS2 knockout mutants and the wild-type M. tuberculosis strain. Total cell extracts of the wild-type and the pstS1 (S1−) and pstS2 (S2−) knockout mutant strains were probed with anti-PstS1 (HBT12) (2, 8) (blot 1), anti-PstS2 (2A1-2) (17, 19) (blot 2), and anti-PstS3 (2F-8) (7, 17) (blot 3), and goat alkaline phosphatase-conjugated anti-mouse immunoglobulin G (Sigma). Bound antibodies were detected using BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium visualization solution (Promega). The electrophoretic mobilities of the rainbow-colored protein molecular mass markers (Amersham Pharmacia Biotech) as observed on the blots are indicated on the left.