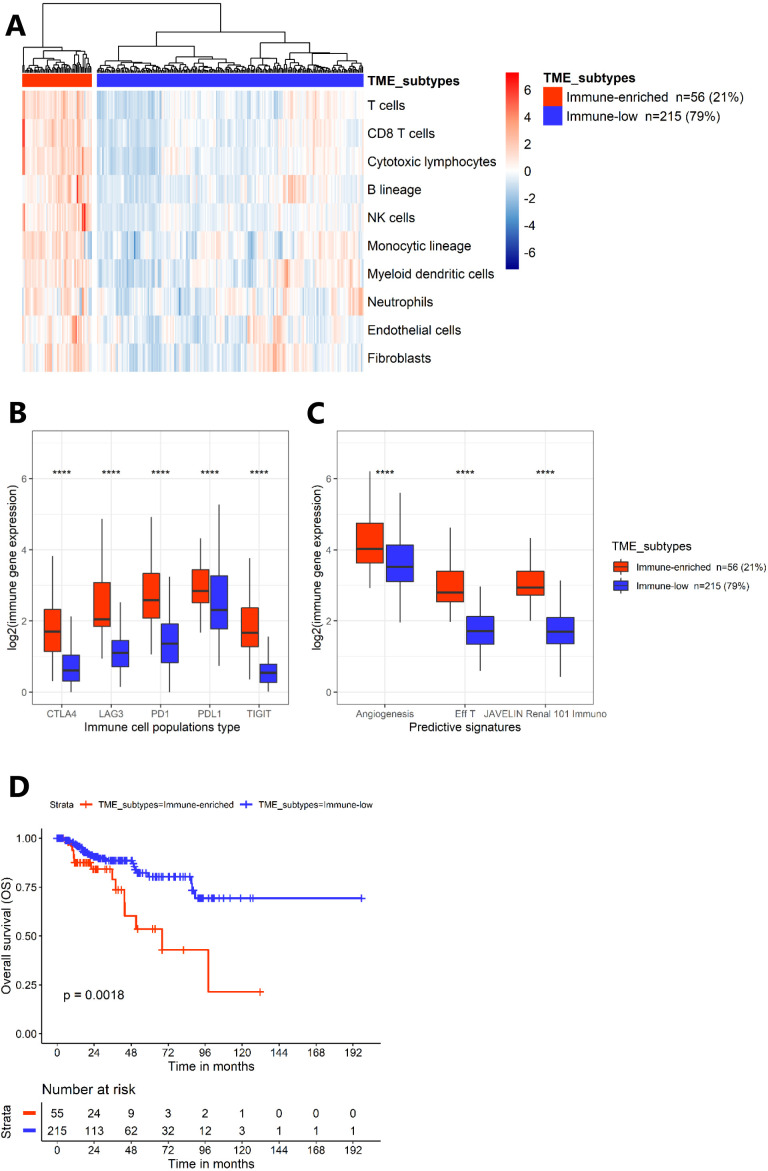
Figure 1.
Transcriptomic analyses from The Cancer Genome Atlas kidney papillary cell carcinoma cohort (n=271). (A) Heatmap representing unsupervised analysis from MCP-counter on normalized transcriptomic expression data (k=2), identifying the TME subtypes. (B) Boxplots representing the transcriptomic expression of five checkpoint markers according to clusters from the MCP-counter analysis. (C) Boxplots representing the three predictive gene signatures of response to treatment described in the clear-cell renal cell carcinoma (angiogenesis, effector T-cell (Eff T), JAVELIN Renal 101 Immuno), according to the clusters from MCP-counter. P values were obtained using the two-sided Mann-Whitney test (***p<0.001). (D) Kaplan-Meier survival curves (overall survival (OS)) according to clusters from MCP-counter analysis. CTLA-4, cytotoxic T-lymphocyte–associated antigen 4; LAG-3, lymphocyte activation gene-3; MCP, Microenvironment Cell Populations; NK, natural killer; PD-1, programmed death 1; PD-L1, programmed death-ligand 1; TIGIT, T cell immunoglobulin and ITIM domain; TME, tumor microenvironment.