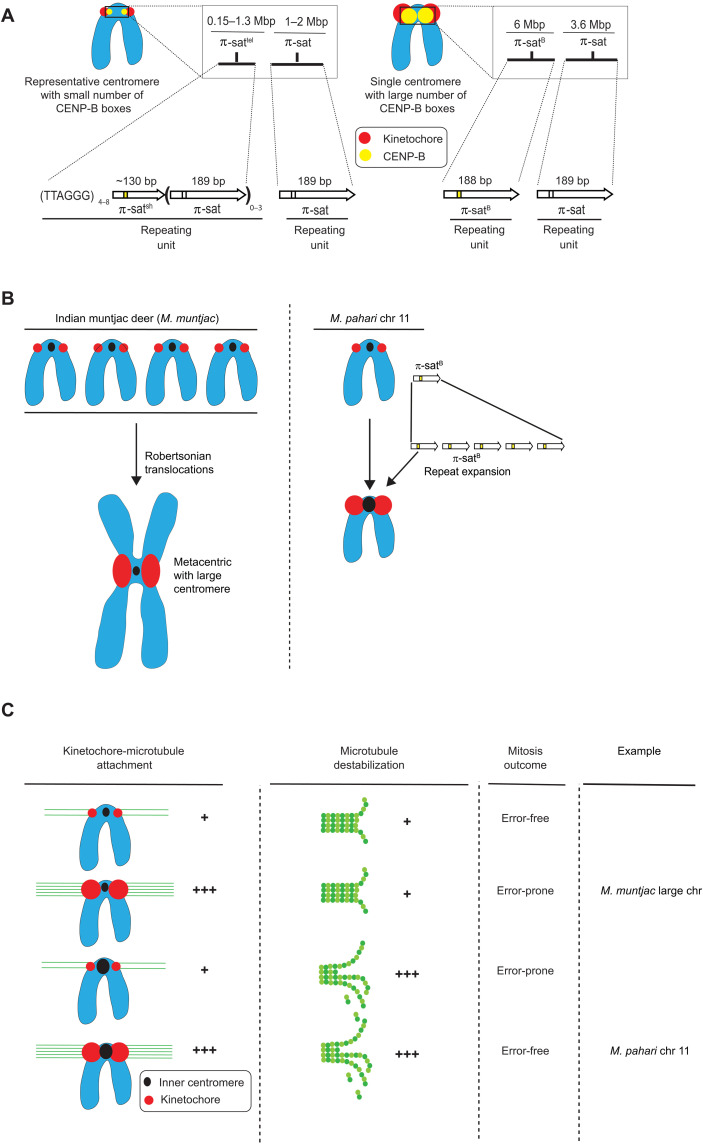
Fig. 8. Divergent centromere DNA, molecular composition, and implications for mitotic chromosome segregation in M. pahari.
(A) Cartoon drawing summarizing the different types of M. pahari centromeres. Most M. pahari centromeres contain a low density of functional CENP-B boxes. Furthermore, these centromeres have two kinds of π-sat. First, the CENP-A–containing region is a stretch of repeating units of π-sat that is short (~130 bp) or long (189 bp) and interspersed with telomeric repeats. This is adjacent to a longer stretch of repeating units of 189-bp π-sat. The second type of M. pahari centromere has a high density of CENP-B boxes and is only found on chromosome 11. This centromere consists of 6 Mbp of homogeneous π-satB. The higher homogeneity of this centromeric DNA suggests that it is evolutionarily more recent relative to the other M. pahari centromeres. (B) A summary of the distinct mechanisms by which Indian munjtac and chromosome 11 from M. pahari centromeres likely became large repetitive arrays observed in the present day. (C) Model to understand different possible outcomes of centromere innovations during mitosis. The typical centromere has relatively low numbers of kinetochore attachments and relatively low amounts of microtubule destabilizer. These two factors balance each other, allowing normal segregation during mitosis. If either pro- or anti-microtubule binding factors are increased in the absence of the other, there will be an imbalance resulting in incorrect segregation during mitosis. Chromosome 11 has higher levels of microtubule destabilizer and more microtubule attachments, but because both factors are increased together, the chromosomes can still undergo error-free mitosis. Large Indian muntjac centromeres, on the other hand, have too strong pro-microtubule binding forces, compromising chromosome segregation (50).