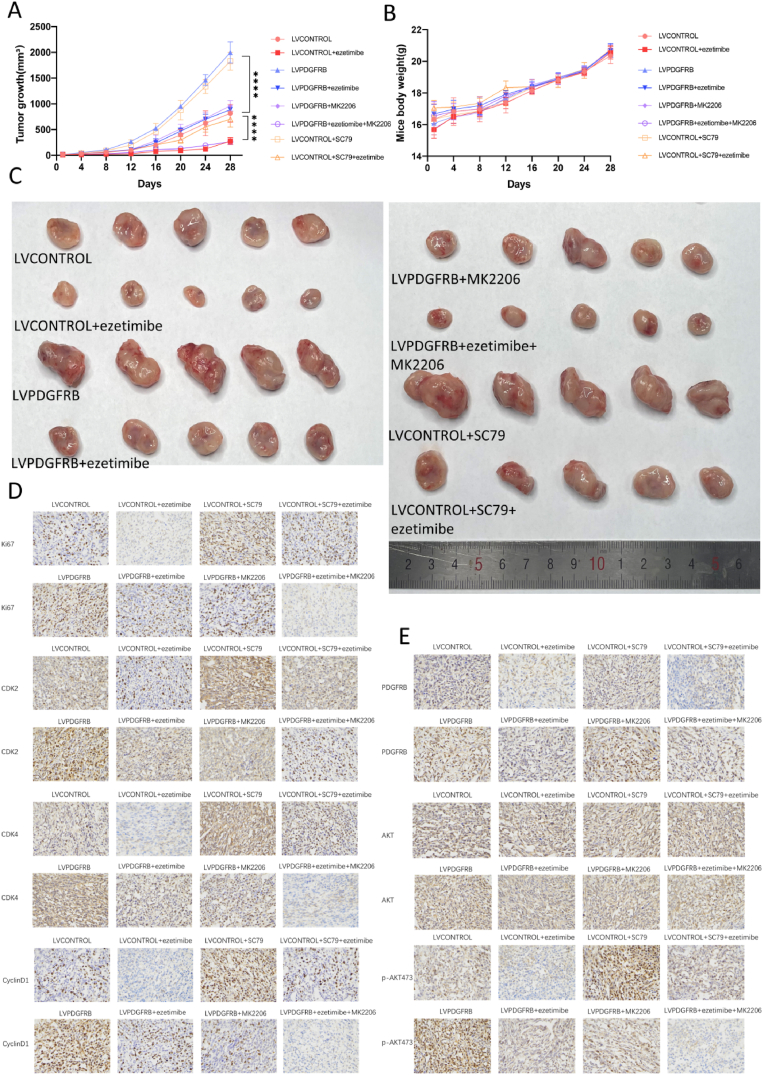
Fig. 6.
The effects of Ezetimibe on TNBC growth and the PDGFRβ/AKT pathway in vivo. (A) MDA-MB-231 cells overexpressing PDGFRβ or transfected with empty vector were separately used to establish subcutaneous breast tumor models in mice. When the tumors reached a size of 5 mm in length, the mice were randomly divided into 8 groups, with 5 mice in each group. The groups received the following treatments: LVCONTROL (empty vector MDA-MB-231) with vehicle treatment, LVCONTROL + ezetimibe (empty vector MDA-MB-231) treated with ezetimibe alone, LVPDGFRB (PDGFRβ-overexpressing MDA-MB-231) with vehicle treatment, LVPDGFRB + ezetimibe (PDGFRβ-overexpressing MDA-MB-231) treated with ezetimibe alone, LVPDGFRB + MK2206 (PDGFRβ-overexpressing MDA-MB-231) treated with MK2206 alone, LVPDGFRB + ezetimibe + MK2206 (PDGFRβ-overexpressing MDA-MB-231) treated with the combination of ezetimibe and MK2206, LVCONTROL + SC79 (empty vector MDA-MB-231) treated with SC79 alone, and LVCONTROL + ezetimibe + SC79 (empty vector MDA-MB-231) treated with the combination of ezetimibe and SC79. During the treatment process, the body weight (A) and tumor length and width (B) were measured at regular intervals. The tumor volume was calculated using the formula: length × width2/2. (C) When the tumor volume reached 2000 mm3, the mice were euthanized while under anesthesia, and the tumors were harvested and photographed. (D) Tumors from xenograft mice with breast cancer were harvested when they reached a size of 2000 mm3. Immunohistochemical staining was performed on the xenograft tumors from each group to detect the expression levels of Ki67, CDK2, CDK4, and CyclinD1. Representative images were captured under a microscope at a magnification of × 400. (E) Immunohistochemical staining was performed on the xenograft tumors from each group to detect the expression levels of PDGFRβ, AKT, and p-AKT473. Representative images were captured under a microscope at a magnification of × 400. The results presented in the figures are representative of three independent experiments and are expressed as mean ± standard deviation (Mean ± SD): *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.