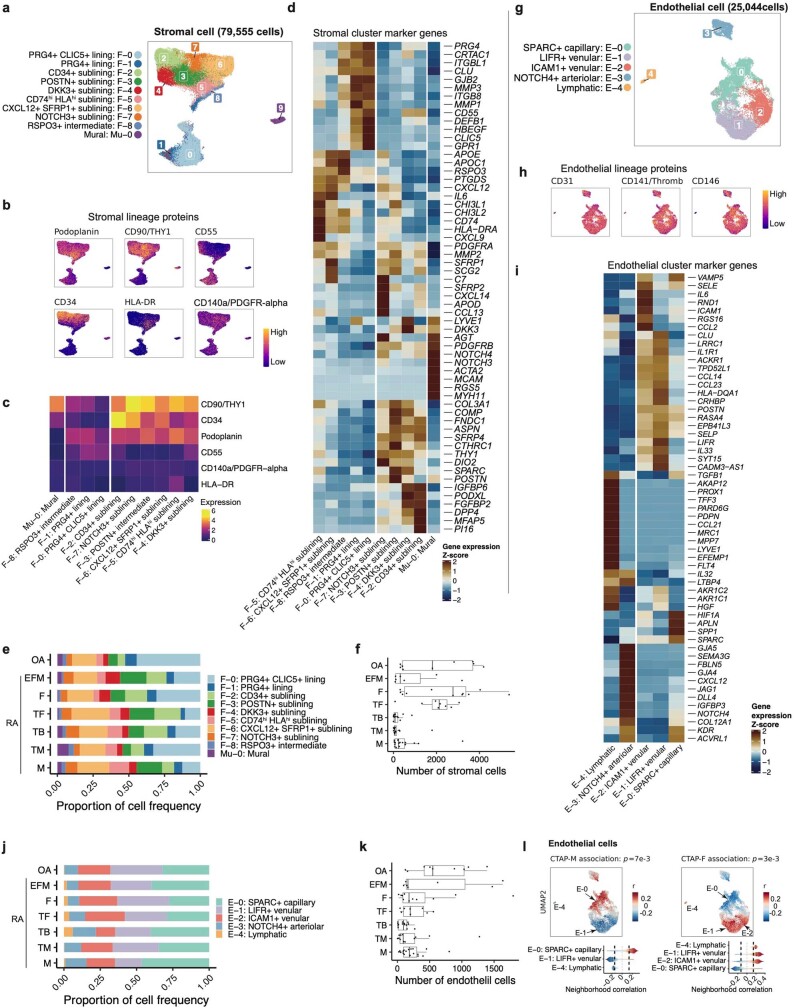
Extended Data Fig. 7. Stromal- and endothelial-specific analysis.
a, Stromal cell UMAP colored by fine-grained cell state clusters, b, Expression of selected surface proteins among stromal cells colored from blue (low) to yellow (high), c, Heatmap of surface protein expression in stromal cell clusters colored according to the average normalized expression across cells in the cluster, d, Heatmap of gene expression in stromal cell clusters colored according to the average normalized expression across cells in the cluster, scaled for each gene across clusters, e, Distribution of stromal cells across clusters, stratified by CTAP. The size of each segment of each bar corresponds to the average proportion of cells in that cluster across donors from that CTAP, f, Number of stromal cells per individual, stratified by CTAP. Points represent individuals (N = 82); OA (N = 9), (EFM (N = 7), F (N = 11), TF (N = 8), TB (N = 14), TM (N = 12), M (N = 18). Box plots show median (vertical bar), 25th and 75th percentiles (lower and upper bounds of the box, respectively) and 1.5 x IQR (or minimum/maximum values; end of whiskers), g, Endothelial cell UMAP colored by fine-grained cell state clusters, h, Expression of selected surface proteins among endothelial cells colored from blue (low) to yellow (high), i, Heatmap of gene expression in endothelial cell clusters colored according to the average normalized expression across cells in the cluster, scaled for each gene across clusters, j, Distribution of endothelial cells across clusters, stratified by CTAP. The size of each segment of each bar corresponds to the average proportion of cells in that cluster across donors from that CTAP. k, Number of endothelial cells per individual, stratified by CTAP. Points represent individuals (N = 82); OA (N = 9), EFM (N = 7), F (N = 11), TF (N = 8), TB (N = 14), TM (n = 12), M (N = 18). Box plots show median (vertical bar), 25th and 75th percentiles (lower and upper bounds of the box, respectively) and 1.5 x IQR (or minimum/maximum values; end of whiskers). l, Association of endothelial cell neighborhoods with CTAP-M and CTAP-F. For these CNA results, cells in UMAPs are colored in red (positive) or blue (negative) if their neighborhood is significantly associated with the CTAP (FDR < 0.05), and gray otherwise. Distributions of neighborhood correlations are shown for clusters with >50% of neighborhoods correlated with the CTAP at FDR < 0.05; global p-values were obtained based on the permutation testing from the CNA package.