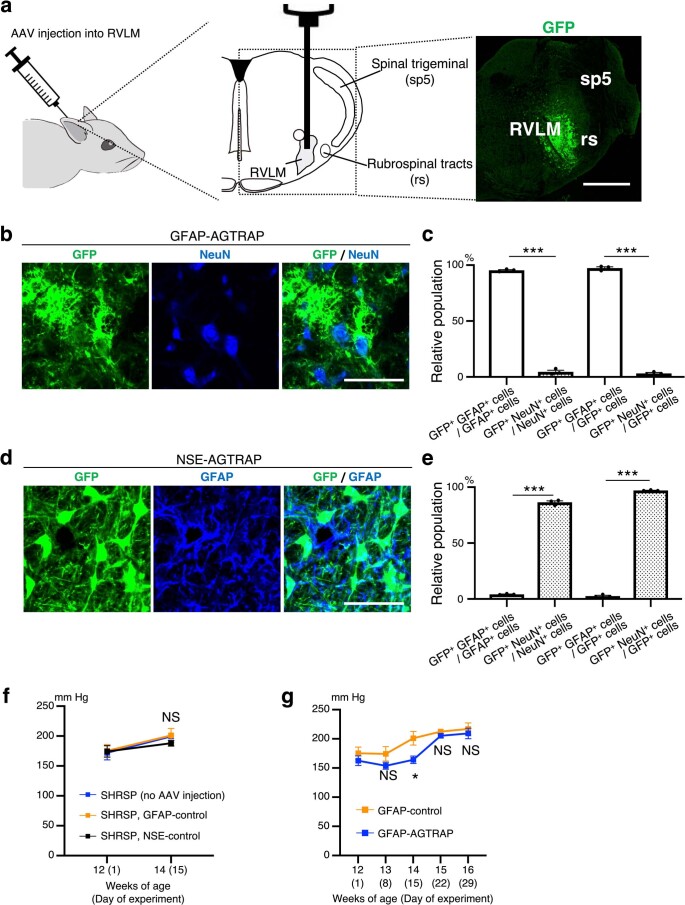
Extended Data Fig. 5. AAV-mediated transduction of RVLM astrocytes or neurons in SHRSPs.
a, Schematic representation of the injection of AAV9 vectors to the RVLM. Micrographic image is representative of three rats analysed in Fig. 3b. GFP-derived fluorescence indicates cells expressing the transgene. Scale bar, 1 mm. b,c, Efficiency and specificity of astrocyte-specific expression of the transgene. (b) Micrographic images of GFP (green) and anti-NeuN immunostaining for mature neurons (blue) of the RVLM of SHRSPs analysed in Fig. 3b. Scale bar, 50 μm. Images are representative of three rats. (c) Relative populations (%) of GFP/ GFAP/ double positive (GFP+ GFAP+) or GFP/ NeuN/ double positive (GFP+ NeuN+) cells were calculated by referring their numbers to those of GFP+, GFAP+ or NeuN+ cells (n = 3 rats for each group). d,e, Efficiency and specificity of neuron-specific expression of the transgene. (d) Micrographic images of GFP (green) and anti-GFAP immunostaining (blue) of the RVLM of SHRSPs analysed in Fig. 3c. Scale bar, 50 μm. Images are representative of three rats. (e) Efficiency and specificity quantified as in (c). f, MAP in SHRSPs injected with the control vectors. Blood pressure was measured and MAP was quantified as in Fig. 1b (n = 6 rats for GFAP-control; n = 7 rats for NSE-control). The data for blue line are identical to those demonstrated with blue line in Extended Data Fig. 1a. g, Four-week time courses of MAP. The experimental protocol was identical to that for Fig. 3 except for the observation period after the AAV injection. The data for Day 1 and Day 15 are identical to those for GFAP-control and GFAP-AGTRAP shown in Fig. 3d,e (GFAP-control: n = 6 rats for Day 1, Day 8, and Day 15; n = 3 rats for Day 22 and Day 29. GFAP-AGTRAP: n = 7 rats for Day 1, Day 8, and Day 15; n = 3 rats for Day 22 and Day 29). Data are presented as mean ± s.e.m. *P < 0.05; ***P < 0.001; NS, not significant; unpaired two-tailed Student’s t-test (c,e) and two-way repeated measures ANOVA with Tukey’s (f) or Bonferroni’s (g) post hoc multiple comparisons test. Details of statistical analyses are provided in Supplementary Information.